7AXZ
 
 | | Ku70/80 complex apo form | | Descriptor: | X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Hnizda, A, Tesina, P, Novak, P, Blundell, T.L. | | Deposit date: | 2020-11-10 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SAP domain forms a flexible part of DNA aperture in Ku70/80.
Febs J., 288, 2021
|
|
5L4Z
 
 | |
5L50
 
 | |
5OPM
 
 | | Crystal structure of D52N/R238W cN-II mutant bound to dATP and free phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPK
 
 | | Crystal structure of D52N/R367Q cN-II mutant bound to dATP and free phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPP
 
 | | Crystal structure of S408R cN-II mutant | | Descriptor: | BROMIDE ION, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPN
 
 | | Crystal structure of R39Q cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPO
 
 | | Crystal structure of R238G cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, MAGNESIUM ION | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5K7Y
 
 | | Crystal structure of enzyme in purine metabolism | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL | | Authors: | Skerlova, J, Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2016-05-27 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interface modulation causes misregulation of purine 5 -nucleotidase in relapsed leukemia.
Bmc Biol., 14, 2016
|
|
5OPL
 
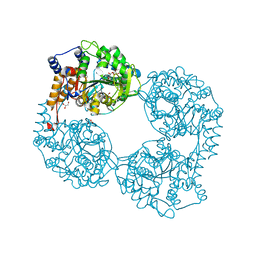 | | Crystal structure of K25E cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, MAGNESIUM ION | | Authors: | Kugler, M, Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
6ZH8
 
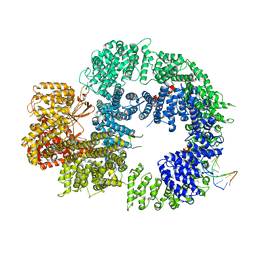 | | Cryo-EM structure of DNA-PKcs:DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZFP
 
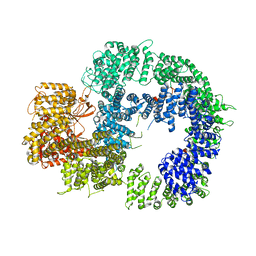 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
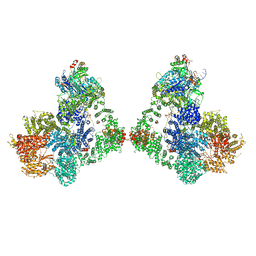 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
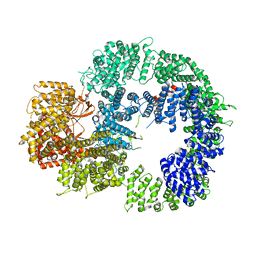 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
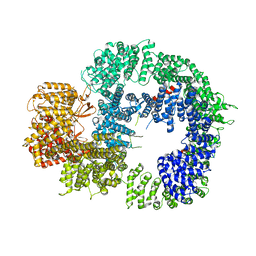 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
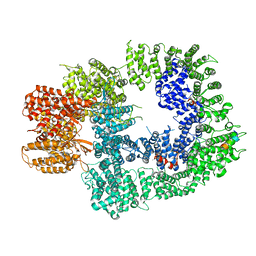 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHA
 
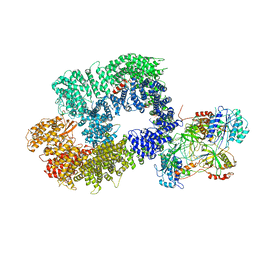 | | Cryo-EM structure of DNA-PK monomer | | Descriptor: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4MWO
 
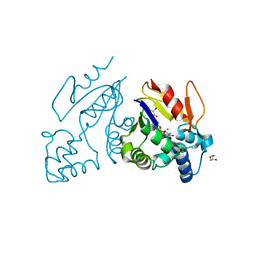 | | Crystal structure of human mitochondrial 5'(3')-deoxyribonucleotidase in complex with the inhibitor CPB-T | | Descriptor: | 1-{3,5-O-[(4-carboxyphenyl)(phosphono)methylidene]-2-deoxy-beta-D-threo-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, 5'(3')-deoxyribonucleotidase, mitochondrial, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2013-09-25 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformationally constrained nucleoside phosphonic acids - potent inhibitors of human mitochondrial and cytosolic 5'(3')-nucleotidases.
Org.Biomol.Chem., 12, 2014
|
|
4L6C
 
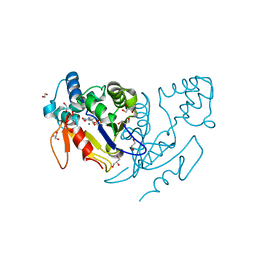 | | Crystal structure of human mitochondrial deoxyribonucleotidase in complex with the inhibitor pib-t | | Descriptor: | 1,2-ETHANEDIOL, 1-{2-deoxy-3,5-O-[(4-iodophenyl)(phosphono)methylidene]-beta-D-threo-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, 5'(3')-deoxyribonucleotidase, ... | | Authors: | Pachl, P, Brynda, J, Rezacova, P. | | Deposit date: | 2013-06-12 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally constrained nucleoside phosphonic acids - potent inhibitors of human mitochondrial and cytosolic 5'(3')-nucleotidases.
Org.Biomol.Chem., 12, 2014
|
|
4PVV
 
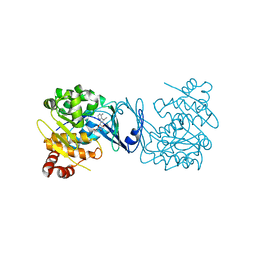 | | Micobacterial Adenosine Kinase in complex with inhibitor | | Descriptor: | 5-ethynyl-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Adenosine kinase | | Authors: | Pichova, I, Hocek, M, Dostal, J, Rezacova, P. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
4O1G
 
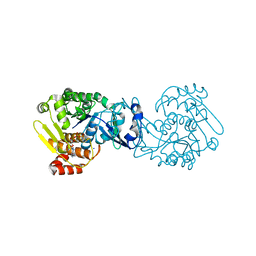 | | MTB adenosine kinase in complex with gamma-Thio-ATP | | Descriptor: | Adenosine kinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SODIUM ION | | Authors: | Dostal, J, Brynda, J, Hocek, M, Pichova, I. | | Deposit date: | 2013-12-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
4O1L
 
 | | Human Adenosine Kinase in complex with inhibitor | | Descriptor: | 5-ethynyl-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Adenosine kinase, MAGNESIUM ION | | Authors: | Brynda, J, Dostal, J, Pichova, I, Hodcek, M. | | Deposit date: | 2013-12-16 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
4NFL
 
 | | Crystal structure of human mitochondrial 5'(3')-deoxyribonucleotidase in complex with the inhibitor NPB-T | | Descriptor: | 1-{2-deoxy-3,5-O-[(4-nitrophenyl)(phosphono)methylidene]-beta-D-threo-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'(3')-deoxyribonucleotidase, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2013-10-31 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.375 Å) | | Cite: | Conformationally constrained nucleoside phosphonic acids - potent inhibitors of human mitochondrial and cytosolic 5'(3')-nucleotidases.
Org.Biomol.Chem., 12, 2014
|
|
