1SK5
 
 | |
1JB8
 
 | | The Crystal Structure of an RNA/DNA Hybrid Reveals Novel Intermolecular Intercalation | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3' | | Authors: | Han, G.W, Kopka, M.L, Langs, D, Dickerson, R.E. | | Deposit date: | 2001-06-02 | | Release date: | 2003-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of an RNADNA hybrid reveals intermolecular
intercalation: Dimer formation by base-pair swapping
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
307D
 
 | | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3') | | Authors: | Han, G.W, Kopka, M.L, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1997-01-07 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis.
J.Mol.Biol., 269, 1997
|
|
1G4Q
 
 | | RNA/DNA HYBRID DECAMER OF CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', ... | | Authors: | Han, G.W. | | Deposit date: | 2000-10-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct-methods determination of an RNA/DNA hybrid decamer at 1.15 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1R75
 
 | |
1FK2
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH MYRISTIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, MYRISTIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK7
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH RICINOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN, RICINOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK3
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK0
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-08 | | Release date: | 2001-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK5
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH OLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, OLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK4
 
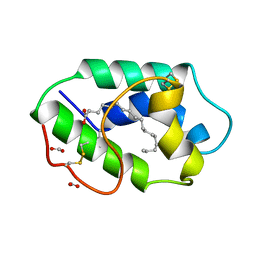 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH STEARIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, STEARIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK1
 
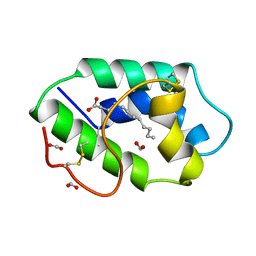 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH LAURIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, LAURIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK6
 
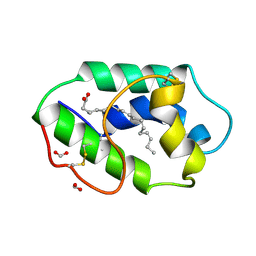 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH ALPHA-LINOLENIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | ALPHA-LINOLENIC ACID, FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
3KSV
 
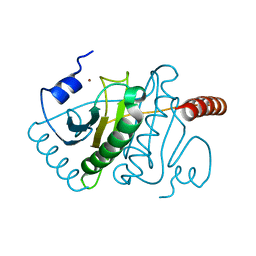 | |
3KPA
 
 | |
2CEP
 
 | |
4L6R
 
 | | Structure of the class B human glucagon G protein coupled receptor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Soluble cytochrome b562 and Glucagon receptor chimera | | Authors: | Siu, F.Y, He, M, de Graaf, C, Han, G.W, Yang, D, Zhang, Z, Zhou, C, Xu, Q, Wacker, D, Joseph, J.S, Liu, W, Lau, J, Cherezov, V, Katritch, V, Wang, M.W, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the human glucagon class B G-protein-coupled receptor.
Nature, 499, 2013
|
|
5XEZ
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
5DHH
 
 | | The crystal structure of nociceptin/orphanin FQ peptide receptor (NOP) in complex with SB-612111 (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5S,7S)-7-{[4-(2,6-dichlorophenyl)piperidin-1-yl]methyl}-1-methyl-6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-ol, OLEIC ACID, ... | | Authors: | Miller, R.L, Thompson, A.A, Trapella, C, Guerrini, R, Malfacini, D, Patel, N, Han, G.W, Cherezov, V, Calo, G, Katritch, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor.
Structure, 23, 2015
|
|
5XR8
 
 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
6ME2
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6LI1
 
 | | Crystal structure of GPR52 ligand free form with flavodoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Flavodoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI2
 
 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
4XNW
 
 | | The human P2Y1 receptor in complex with MRS2500 | | Descriptor: | P2Y purinoceptor 1,Rubredoxin,P2Y purinoceptor 1, ZINC ION, [(1R,2S,4S,5S)-4-[2-iodo-6-(methylamino)-9H-purin-9-yl]-2-(phosphonooxy)bicyclo[3.1.0]hex-1-yl]methyl dihydrogen phosphate | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
