5Z7L
 
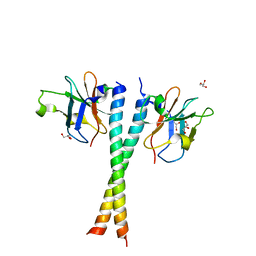 | | Crystal structure of NDP52 SKICH region in complex with NAP1 | | Descriptor: | 5-azacytidine-induced protein 2, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL | | Authors: | Fu, T, Pan, L.F. | | Deposit date: | 2018-01-29 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7EA7
 
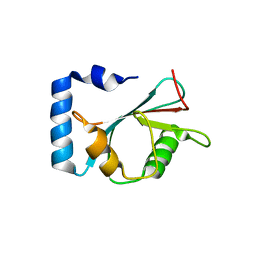 | | crystal structure of NAP1 LIR in complex with GABARAP | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, NAP1_LIR motif | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
7EA2
 
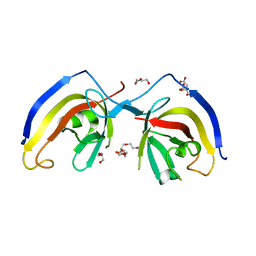 | | crystal structure of NAP1 FIR in complex with RB1CC1 Claw domain | | Descriptor: | 5-azacytidine-induced protein 2,RB1-inducible coiled-coil protein 1, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
7EAA
 
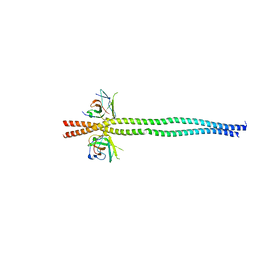 | | crystal structure of NDP52 SKICH domain in complex with RB1CC1 coiled-coil domain | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2, RB1-inducible coiled-coil protein 1 | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
5Z7A
 
 | | Crystal structure of NDP52 SKICH region | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q. | | Deposit date: | 2018-01-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Z7G
 
 | | Crystal structure of TAX1BP1 SKICH region in complex with NAP1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Tax1-binding protein 1 | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q, Wang, Y.L, Hu, S.C. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8GH8
 
 | | RuvA Holliday junction DNA complex | | Descriptor: | DNA (34-MER), Holliday junction branch migration complex subunit RuvA | | Authors: | Rish, A.D, Fu, T. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | RuvA Holliday junction DNA complex
To Be Published
|
|
5GP7
 
 | | Structural basis for the binding between Tankyrase-1 and USP25 | | Descriptor: | GLYCEROL, Tankyrase-1, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Liu, J, Xu, D, Fu, T, Pan, L. | | Deposit date: | 2016-08-01 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | USP25 regulates Wnt signaling by controlling the stability of tankyrases
Genes Dev., 31, 2017
|
|
6JV5
 
 | | Crystal structure of 5-methylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(5CM)P*GP*CP*TP*GP*G)-3') | | Authors: | Zhang, L, Wang, Y.X. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5JQE
 
 | | Crystal structure of caspase8 tDED | | Descriptor: | Sugar ABC transporter substrate-binding protein,Caspase-8 chimera | | Authors: | Fu, T, Li, Y, Lu, A, Wu, H. | | Deposit date: | 2016-05-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.157 Å) | | Cite: | Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex.
Mol. Cell, 64, 2016
|
|
5ZAT
 
 | | Crystal structure of 5-carboxylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(CAC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5ZAS
 
 | | Crystal structure of 5-formylcytosine containing decamer dsDNA | | Descriptor: | BICARBONATE ION, DNA (5'-D(*CP*CP*AP*GP*(5FC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
6JV3
 
 | |
2NA6
 
 | | Transmembrane domain of mouse Fas/CD95 death receptor | | Descriptor: | Tumor necrosis factor receptor superfamily member 6 | | Authors: | Fu, Q, Chou, J.J, Wu, H, Fu, T, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis and Functional Role of Intramembrane Trimerization of the Fas/CD95 Death Receptor.
Mol.Cell, 61, 2016
|
|
6NHW
 
 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6XKJ
 
 | | Cryo-EM structure of CARD8-CARD filament | | Descriptor: | Caspase recruitment domain-containing protein 8 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | Deposit date: | 2020-06-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
6XKK
 
 | | Cryo-EM structure of the NLRP1-CARD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | Deposit date: | 2020-06-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
6N1H
 
 | | Cryo-EM structure of ASC-CARD filament | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N1I
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7BV6
 
 | | Crystal structure of the autophagic STX17/SNAP29/VAMP8 SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BV4
 
 | | Crystal structure of STX17 LIR region in complex with GABARAP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4O7Q
 
 | |
4ZJB
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) in complex with holo-ACP from Helicobacter pylori | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Zhang, L, Zhang, L, Shen, X, Jiang, H. | | Deposit date: | 2015-04-29 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of FabZ-ACP complex reveals a dynamic seesaw-like catalytic mechanism of dehydratase in fatty acid biosynthesis.
Cell Res., 26, 2016
|
|
5X0W
 
 | | Molecular mechanism for the binding between Sharpin and HOIP | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Sharpin | | Authors: | Liu, J, Li, F, Cheng, X, Pan, L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into SHARPIN-Mediated Activation of HOIP for the Linear Ubiquitin Chain Assembly
Cell Rep, 21, 2017
|
|
