6LUB
 
 | | Crystal Structure of EGFR(L858R/T790M/C797S) in complex with CH7233163 | | Descriptor: | Epidermal growth factor receptor, N-[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]-7-(4-methylpiperazin-1-yl)-5-propan-2-yl-9-[2,2,2-tris(fluoranyl)ethoxy]pyrido[4,3-b]indol-3-amine | | Authors: | Kawauchi, H, Fukami, T.A, Sato, S, Endo, M, Torizawa, T, Kashima, K, Chiba, T, Sakamoto, H. | | Deposit date: | 2020-01-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Mol.Cancer Ther., 19, 2020
|
|
6LUD
 
 | | Crystal Structure of EGFR(L858R/T790M/C797S) in complex with Osimertinib | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Kawauchi, H, Fukami, T.A, Sato, S, Endo, M, Torizawa, T, Kashima, K, Chiba, T, Sakamoto, H. | | Deposit date: | 2020-01-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Mol.Cancer Ther., 19, 2020
|
|
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UMH
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | F-box only protein 2, NICKEL (II) ION | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
6KX0
 
 | | Crystal structure of SN-101 mAb non-liganded form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
6KX1
 
 | | Crystal structure of SN-101 mAb in complex with MUC1 glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain, ... | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
3VW9
 
 | | Human Glyoxalase I with an N-hydroxypyridone inhibitor | | Descriptor: | 1-hydroxy-6-[1-(3-methoxypropyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-4-phenylpyridin-2(1H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lactoylglutathione lyase, ... | | Authors: | Fukami, T.A, Irie, M, Matsuura, T. | | Deposit date: | 2012-08-10 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Design and evaluation of azaindole-substituted N-hydroxypyridones as glyoxalase I inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2RJ2
 
 | | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution | | Descriptor: | CHLORIDE ION, F-box only protein 2, NICKEL (II) ION | | Authors: | Vaijayanthimala, S, Velmurugan, D, Mizushima, T, Yamane, T, Yoshida, Y, Tanaka, K. | | Deposit date: | 2007-10-14 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution
To be Published
|
|
3A4P
 
 | | human c-MET kinase domain complexed with 6-benzyloxyquinoline inhibitor | | Descriptor: | (2E)-3-{6-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]quinolin-3-yl}-N-methylprop-2-enamide, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Fukami, T.A, Kadono, S, Yamamuro, M, Matsuura, T. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1D9N
 
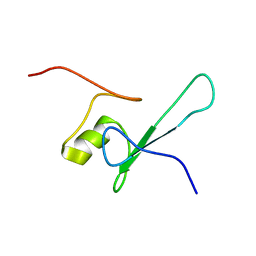 | | SOLUTION STRUCTURE OF THE METHYL-CPG-BINDING DOMAIN OF THE METHYLATION-DEPENDENT TRANSCRIPTIONAL REPRESSOR MBD1/PCM1 | | Descriptor: | METHYL-CPG-BINDING PROTEIN MBD1 | | Authors: | Ohki, I, Shimotake, N, Fujita, N, Nakao, M, Shirakawa, M. | | Deposit date: | 1999-10-28 | | Release date: | 2000-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1.
EMBO J., 18, 1999
|
|
7YUZ
 
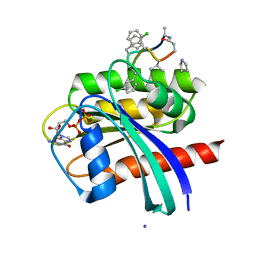 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP8784 | | Descriptor: | AP8784, GUANOSINE-5'-DIPHOSPHATE, IODIDE ION, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
7YV1
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor LUNA18 and KA30L Fab | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, KA30L Fab H-chain, ... | | Authors: | Irie, M, Fukami, T.A, Matsuo, A, Saka, K, Nishimura, M, Saito, H, Torizawa, T, Tanada, M, Ohta, A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
1I2H
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSD-ZIP45(HOMER1C/VESL-1L)CONSERVED HOMER 1 DOMAIN | | Descriptor: | PSD-ZIP45(HOMER-1C/VESL-1L) | | Authors: | Irie, K, Nakatsu, T, Mitsuoka, K, Fujiyoshi, Y, Kato, H. | | Deposit date: | 2001-02-09 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Homer 1 Family Conserved Region Reveals the Interaction Between the EVH1 Domain and
Own Proline-rich Motif
J.Mol.Biol., 318, 2002
|
|
5GXQ
 
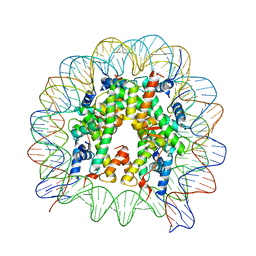 | | The crystal structure of the nucleosome containing H3.6 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Taguchi, H, Xie, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
5X7X
 
 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.184 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
7DBH
 
 | | The mouse nucleosome structure containing H3mm18 | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2020-10-20 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
5B71
 
 | | Crystal structure of complement C5 in complex with SKY59 | | Descriptor: | Complement C5 beta chain, SKY59 Fab heavy chain, SKY59 Fab light chain | | Authors: | Irie, M, Shimizu, Y, Sampei, Z, Fukuzawa, T. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases.
Sci Rep, 7, 2017
|
|
5B1M
 
 | | The mouse nucleosome structure containing H3.1 | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B type 3-A, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
5B1L
 
 | | The mouse nucleosome structure containing H3t | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
7VBM
 
 | | The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2021-08-31 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
