1LNQ
 
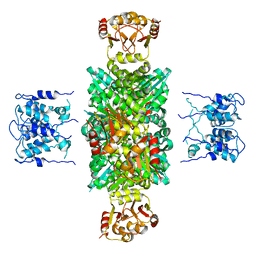 | | CRYSTAL STRUCTURE OF MTHK AT 3.3 A | | Descriptor: | CALCIUM ION, POTASSIUM CHANNEL RELATED PROTEIN | | Authors: | Jiang, Y, Lee, A, Chen, J, Cadene, M, Chait, B.T, Mackinnon, R. | | Deposit date: | 2002-05-03 | | Release date: | 2002-06-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | CRYSTAL STRUCTURE AND MECHANISM OF A CALCIUM-GATED POTASSIUM CHANNEL
Nature, 417, 2002
|
|
1TAF
 
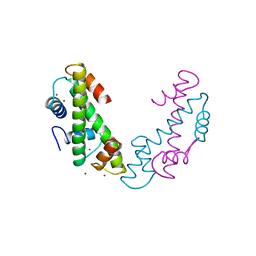 | | DROSOPHILA TBP ASSOCIATED FACTORS DTAFII42/DTAFII62 HETEROTETRAMER | | Descriptor: | TFIID TBP ASSOCIATED FACTOR 42, TFIID TBP ASSOCIATED FACTOR 62, ZINC ION | | Authors: | Xie, X, Kokubo, T, Cohen, S.L, Mirza, U.A, Hoffmann, A, Chait, B.T, Roeder, R.G, Nakatani, Y, Burley, S.K. | | Deposit date: | 1996-06-01 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural similarity between TAFs and the heterotetrameric core of the histone octamer.
Nature, 380, 1996
|
|
1ORQ
 
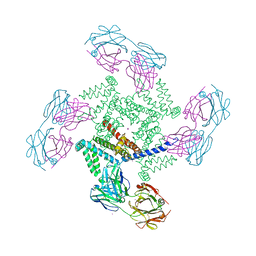 | | X-ray structure of a voltage-dependent potassium channel in complex with an Fab | | Descriptor: | 6E1 Fab heavy chain, 6E1 Fab light chain, CADMIUM ION, ... | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
1ORS
 
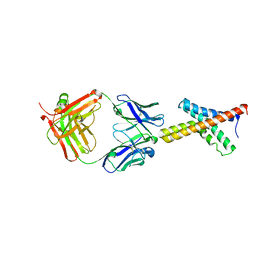 | | X-ray structure of the KvAP potassium channel voltage sensor in complex with an Fab | | Descriptor: | 33H1 Fab heavy chain, 33H1 Fab light chain, potassium channel | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
1KPK
 
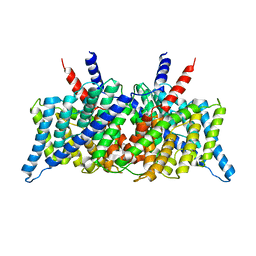 | | Crystal Structure of the ClC Chloride Channel from E. coli | | Descriptor: | putative channel transporter | | Authors: | Dutzler, R, Campbell, E.B, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-12-31 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of
anion selectivity.
Nature, 415, 2002
|
|
1ID1
 
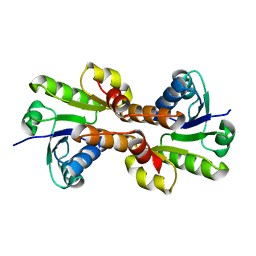 | | CRYSTAL STRUCTURE OF THE RCK DOMAIN FROM E.COLI POTASSIUM CHANNEL | | Descriptor: | PUTATIVE POTASSIUM CHANNEL PROTEIN | | Authors: | Jiang, Y, Pico, A, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel.
Neuron, 29, 2001
|
|
1KPL
 
 | | Crystal Structure of the ClC Chloride Channel from S. typhimurium | | Descriptor: | CHLORIDE ION, N-OCTANE, PENTADECANE, ... | | Authors: | Dutzler, R, Campbell, E.B, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-12-31 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of
anion selectivity.
Nature, 415, 2002
|
|
6DRD
 
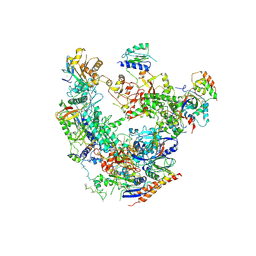 | | RNA Pol II(G) | | Descriptor: | DNA-directed RNA polymerase II subunit GRINL1A, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Yu, X, Jishage, M, Shi, Y, Ganesan, S, Sali, A, Chait, B.T, Asturias, F, Roeder, R.G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-12 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1BL8
 
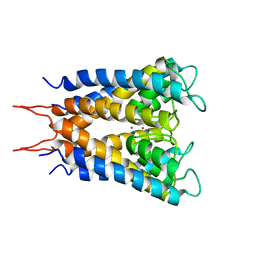 | | POTASSIUM CHANNEL (KCSA) FROM STREPTOMYCES LIVIDANS | | Descriptor: | POTASSIUM ION, PROTEIN (POTASSIUM CHANNEL PROTEIN) | | Authors: | Doyle, D.A, Cabral, J.M, Pfuetzner, R.A, Kuo, A, Gulbis, J.M, Cohen, S.L, Chait, B.T, Mackinnon, R. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the potassium channel: molecular basis of K+ conduction and selectivity.
Science, 280, 1998
|
|
2JMI
 
 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
1CMO
 
 | | IMMUNOGLOBULIN MOTIF DNA-RECOGNITION AND HETERODIMERIZATION FOR THE PEBP2/CBF RUNT-DOMAIN | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Nagata, T, Gupta, V, Sorce, D, Kim, W.Y, Sali, A, Chait, B.T, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-11 | | Release date: | 2000-01-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain.
Nat.Struct.Biol., 6, 1999
|
|
2JMJ
 
 | | NMR solution structure of the PHD domain from the yeast YNG1 protein in complex with H3(1-9)K4me3 peptide | | Descriptor: | Histone H3, Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
6V69
 
 | | Structures of GCP4 and GCP5 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 4, Gamma-tubulin complex component 5 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V6C
 
 | | Structure of GCP6 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 6 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V6S
 
 | | Structure of the native human gamma-tubulin ring complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Gamma-tubulin complex component 2, ... | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V5V
 
 | | Structure of gamma-tubulin in the native human gamma-tubulin ring complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Tubulin gamma-1 chain | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V6B
 
 | | Structures of GCP2 and GCP3 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 2, Gamma-tubulin complex component 3 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
7P56
 
 | | Variant Surface Glycoprotein 2 (VSG2, MiTat1.2, VSG221) Bound to Calcium | | Descriptor: | CALCIUM ION, Variant surface glycoprotein MITAT 1.2, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gkeka, A, Aresta-Branco, F, Stebbins, C.E, Papavasiliou, F.N. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | Immunodominant surface epitopes power immune evasion in the African trypanosome.
Cell Rep, 42, 2023
|
|
7P5D
 
 | | Variant Surface Glycoprotein 3 (VSG3, MiTat1.3, VSG224) mutant (serine 317 and 319 to alanine) | | Descriptor: | Variant surface glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gkeka, A, Aresta-Branco, F, Stebbins, C.E, Papavasiliou, F.N. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Immunodominant surface epitopes power immune evasion in the African trypanosome.
Cell Rep, 42, 2023
|
|
7P57
 
 | | VSG2 mutant structure lacking the calcium binding pocket | | Descriptor: | Variant surface glycoprotein MITAT 1.2, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gkeka, A, Aresta-Branco, F, Stebbins, C.E, Papavasiliou, F.N. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Immunodominant surface epitopes power immune evasion in the African trypanosome.
Cell Rep, 42, 2023
|
|
7P59
 
 | | Variant Surface Glycoprotein 3 (VSG3, MiTat1.3, VSG224) with two O-linked post-translational modifications | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gkeka, A, Arest-Branco, F, Stebbins, C.E, Papavasiliou, F.N. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Immunodominant surface epitopes power immune evasion in the African trypanosome.
Cell Rep, 42, 2023
|
|
7P5B
 
 | | Variant Surface Glycoprotein 3 (VSG3, MiTat1.3, VSG224) mutant (serine 319 to alanine), single O-linked glycosylated at Ser317 | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gkeka, A, Aresta-Branco, F, Stebbins, C.E, Papavasiliou, F.N. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Immunodominant surface epitopes power immune evasion in the African trypanosome.
Cell Rep, 42, 2023
|
|
7P5A
 
 | | Variant Surface Glycoprotein 3 (VSG3, MiTat1.3, VSG224) mutant (serine 317 to alanine), single O-linked glycosylated at Ser319 | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gkeka, A, Aresta-Branco, F, Stebbins, C.E, Papavasiliou, F.N. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Immunodominant surface epitopes power immune evasion in the African trypanosome.
Cell Rep, 42, 2023
|
|
6XEZ
 
 | | Structure of SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Llewellyn, E.C, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.
Cell, 182, 2020
|
|
8G0I
 
 | | High Affinity nanobodies against GFP | | Descriptor: | CHLORIDE ION, Green fluorescent protein, LaG24 Nanobody, ... | | Authors: | Ketaren, N.E, Rout, M.P, Almo, S. | | Deposit date: | 2023-01-31 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique Binding and Stabilization Mechanisms Employed By and Engineered Into Nanobodies
Biorxiv, 2023
|
|
