6QAT
 
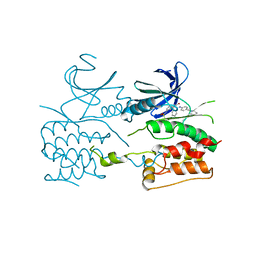 | | Crystal structure of ULK2 in complexed with hesperadin | | Descriptor: | N-{(3Z)-2-oxo-3-[phenyl({4-[(piperidin-1-yl)methyl]phenyl}amino)methylidene]-2,3-dihydro-1H-indol-5-yl}ethanesulfonamide, Serine/threonine-protein kinase ULK2 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6QAS
 
 | | Crystal structure of ULK1 in complexed with PF-03814735 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6QAU
 
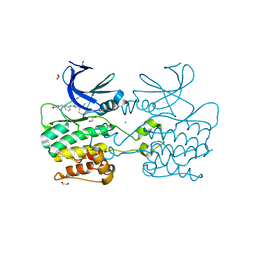 | | Crystal structure of ULK2 in complexed with MRT67307 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6QAV
 
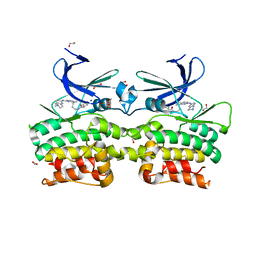 | | Crystal structure of ULK2 in complexed with MRT68921 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SODIUM ION, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
5LXC
 
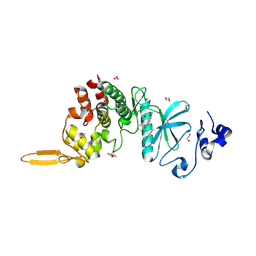 | | Crystal structure of DYRK2 in complex with EHT 5372 (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2,4-dichlorophenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
6QU2
 
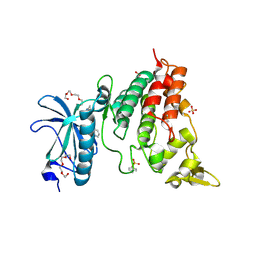 | | Crystal structure of DYRK1A complexed with FC162 inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-cyclopropyl-2-pyridin-3-yl-[1,3]thiazolo[5,4-f]quinazolin-9-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of DYRK1A complexed with FC162 inhibitor
To Be Published
|
|
6RCG
 
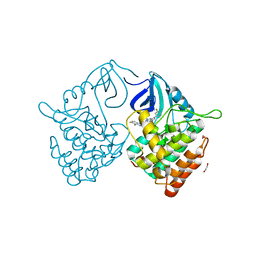 | | Crystal structure of Casein kinase 1 delta (CK1 delta) complexed with SR3029 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, ~{N}-[[6,7-bis(fluoranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(3-fluorophenyl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Roush, W.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Casein kinase 1 delta (CK1 delta) complexed with SR3029 inhibitor
To Be Published
|
|
6RCH
 
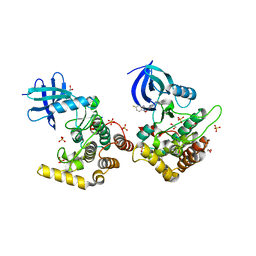 | | Crystal structure of Casein kinase I isoform delta (CK1 delta) complexed with SR4133 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, SODIUM ION, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Roush, W.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of Casein kinase I isoform delta (CK1 delta) complexed with SR4133 inhibitor
To Be Published
|
|
6RCT
 
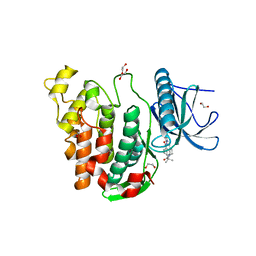 | | Crystal structure of CLK3 in complex with T3-CLK | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-methyl-1-(4-methylpiperazin-1-yl)-1-oxidanylidene-propan-2-yl]-~{N}-(6-pyridin-4-ylimidazo[1,2-a]pyridin-2-yl)benzamide, Dual specificity protein kinase CLK3 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of CLK3 in complex with TP003
To Be Published
|
|
7OPG
 
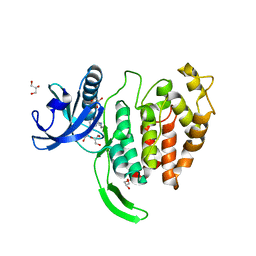 | | Crystal structure of CLK1 in complex with compound 2 (CC513) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[2-(propylamino)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]phenol, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Routier, S, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of CLK1 in complex with compound 2 (CC513)
To Be Published
|
|
5MUF
 
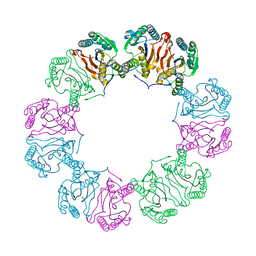 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in its enzymatically active dodecameric form induced by the presence of the N-terminal WDPNWD motif | | Descriptor: | PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
7QRW
 
 | |
7QS4
 
 | |
7QRZ
 
 | |
7QS2
 
 | |
7QS5
 
 | |
7QS3
 
 | |
7QRV
 
 | | Crystal structure of NHL domain of TRIM2 (full C-terminal) | | Descriptor: | 1,2-ETHANEDIOL, Tripartite motif-containing protein 2 | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparative structural analyses of the NHL domains from the human E3 ligase TRIM-NHL family.
Iucrj, 9, 2022
|
|
7QRY
 
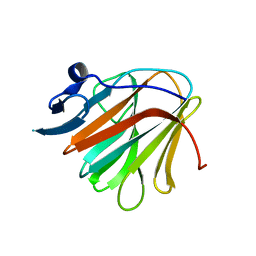 | |
7QS1
 
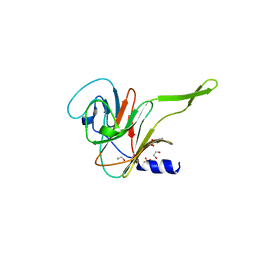 | |
7QRX
 
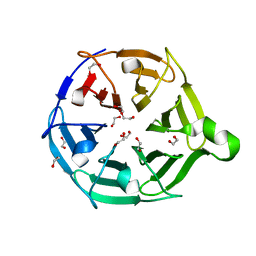 | |
7QS0
 
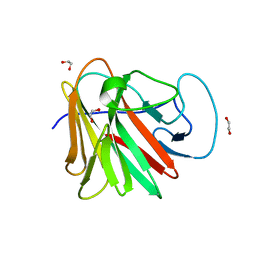 | |
5LXD
 
 | | Crystal structure of DYRK2 in complex with EHT 1610 (compound 2) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
4QSS
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-methylpyrrolidin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4QSR
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) with bound MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATPase family AAA domain-containing protein 2, GLYCEROL, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
