4OVG
 
 | | E. coli sliding clamp in complex with (R)-9-(2-amino-2-oxoethyl)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-9-(2-amino-2-oxoethyl)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4MBI
 
 | | Discovery of Pyrazolo[1,5a]pyrimidine-based Pim1 Inhibitors | | Descriptor: | N,N-dimethyl-N'-[3-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-5-yl]ethane-1,2-diamine, Serine/threonine-protein kinase pim-1 | | Authors: | Azevedo, R, Fischmann, T.O. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of pyrazolo[1,5-a]pyrimidine-based Pim inhibitors: A template-based approach.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4PNV
 
 | |
4N98
 
 | |
4N9A
 
 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
3QBY
 
 | | Crystal structure of the PWWP domain of human Hepatoma-derived growth factor 2 | | Descriptor: | H4K20me3 Histone H4 Peptide, Hepatoma-derived growth factor-related protein 2, SULFATE ION, ... | | Authors: | Zeng, H, Tempel, W, Amaya, M.F, Adams-Cioaba, M.A, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
4RXD
 
 | |
7C96
 
 | | Avr1d:GmPUB13 U-box | | Descriptor: | RING-type E3 ubiquitin transferase, RxLR effector protein Avh6 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4N97
 
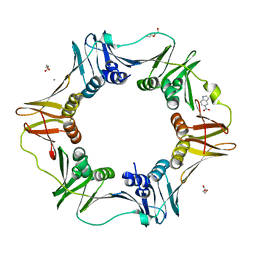 | | E. coli sliding clamp in complex with 5-nitroindole | | Descriptor: | 5-nitro-1H-indole, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N99
 
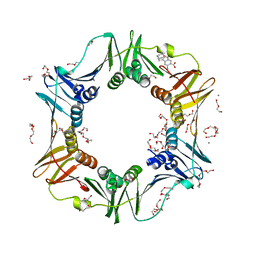 | | E. coli sliding clamp in complex with 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid | | Descriptor: | 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
7VUP
 
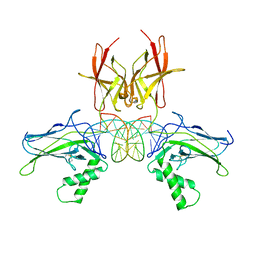 | |
7VUQ
 
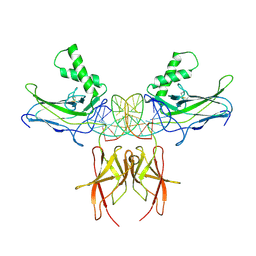 | |
4N95
 
 | | E. coli sliding clamp in complex with 5-chloroindoline-2,3-dione | | Descriptor: | 5-chloro-1H-indole-2,3-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4PNU
 
 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-((R)-1-carboxy-2-phenylethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-(2-{[(1R)-1-carboxy-2-phenylethyl]amino}-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4Q0D
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP, methotrexate and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-L-glutamic acid. | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | Authors: | Kumar, V.P, Anderson, K.S. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.449 Å) | | Cite: | Structural studies provide clues for analog design of specific inhibitors of Cryptosporidium hominis thymidylate synthase-dihydrofolate reductase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N73
 
 | |
4RXE
 
 | |
4RXC
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Homorisedronate BPH-6 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [1-hydroxy-3-(pyridin-3-yl)propane-1,1-diyl]bis(phosphonic acid) | | Authors: | Cao, R, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Farnesyl diphosphate synthase inhibitors with unique ligand-binding geometries.
ACS Med Chem Lett, 6, 2015
|
|
2LK3
 
 | | U2/U6 Helix I | | Descriptor: | RNA (5'-R(*GP*GP*CP*UP*UP*AP*GP*AP*UP*CP*AP*GP*AP*AP*AP*UP*GP*AP*UP*CP*AP*GP*CP*C)-3') | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X.E, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
4N96
 
 | | E. coli sliding clamp in complex with 6-nitroindazole | | Descriptor: | 6-NITROINDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
2LKR
 
 | | Yeast U2/U6 complex | | Descriptor: | RNA (111-MER) | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
7VJL
 
 | | The crystal structure of FGFR4 kinase domain in complex with N-(5-cyano-4-((2-methoxyethyl)amino)pyridin-2-yl)-7-(2,2,2-trifluoroacetyl)-3,4-dihydro-1,8-naphthyridine-1(2H)-carboxamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[5-(aminomethyl)-4-(2-methoxyethylamino)pyridin-2-yl]-7-[2,2,2-tris(fluoranyl)ethanoyl]-3,4-dihydro-2H-1,8-naphthyridine-1-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.900173 Å) | | Cite: | Characterization of an aromatic trifluoromethyl ketone as a new warhead for covalently reversible kinase inhibitor design.
Bioorg.Med.Chem., 50, 2021
|
|
5XSR
 
 | | novel orally efficacious inhibitors complexed with PARP1 | | Descriptor: | 6-fluoranyl-2-(4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)-1~{H}-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-06-15 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of 2-(4,5,6,7-tetrahydrothienopyridin-2-yl)-benzoimidazole carboxamides as novel orally efficacious Poly(ADP-ribose)polymerase (PARP) inhibitors
Eur J Med Chem, 145, 2018
|
|
8HC0
 
 | | Crystal structure of the extracellular domains of GPR110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor F1, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, F.F, Song, G.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Extracellular Domains of GPR110.
J.Mol.Biol., 435, 2023
|
|
5X04
 
 | | 12:0-ACP thioesterase from Umbellularia californica | | Descriptor: | Dodecanoyl-[acyl-carrier-protein] hydrolase, chloroplastic | | Authors: | Xue, S, Feng, Y. | | Deposit date: | 2017-01-19 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insight into Acyl-ACP Thioesterase toward Substrate Specificity Design.
ACS Chem. Biol., 12, 2017
|
|
