7AKJ
 
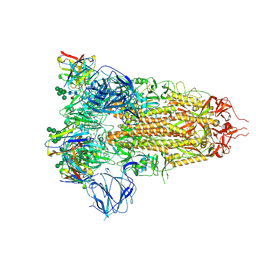 | | Structure of the SARS-CoV spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-10-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
7AKD
 
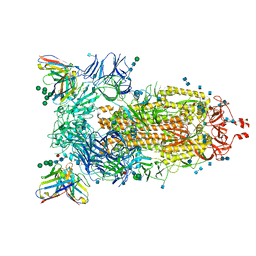 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 47D11 neutralizing antibody heavy chain, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
2YYF
 
 | | Purification and structural characterization of a D-amino acid containing conopeptide, marmophine, from Conus marmoreus | | Descriptor: | M-conotoxin mr12 | | Authors: | Huang, F, Du, W, Han, Y, Wang, C. | | Deposit date: | 2007-04-29 | | Release date: | 2008-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Purification and structural characterization of a D-amino acid-containing conopeptide, conomarphin, from Conus marmoreus.
Febs J., 275, 2008
|
|
5XXH
 
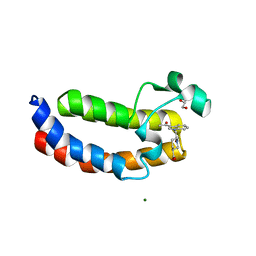 | | Crystal Structure Analysis of the CBP | | Descriptor: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
5Y4E
 
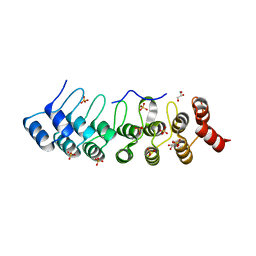 | | Crystal Structure of AnkB Ankyrin Repeats R8-14 in complex with autoinhibition segment AI-b | | Descriptor: | Ankyrin-2,Ankyrin-2, GLYCEROL, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
8I22
 
 | | Acyl-ACP synthetase structure bound to pimelic acid monoethyl ester | | Descriptor: | 7-ethoxy-7-oxidanylidene-heptanoic acid, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase
Nat.Struct.Mol.Biol., 2025
|
|
8I35
 
 | | Acyl-ACP synthetase structure bound to oleic acid | | Descriptor: | Acyl-acyl carrier protein synthetase, OLEIC ACID | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-16 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I3I
 
 | | Acyl-ACP synthetase structure bound to AMP-PNP in the presence of MgCl2 | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-17 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I6M
 
 | | Acyl-ACP synthetase structure bound to AMP-C18:1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, MAGNESIUM ION, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8HZX
 
 | | Acyl-ACP synthetase structure-2 | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I51
 
 | | Acyl-ACP synthetase structure bound to AMP-MC7 | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-21 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I49
 
 | | Acyl-ACP synthetase structure bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I8D
 
 | | Acyl-ACP synthetase structure bound to MC7-ACP | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-07 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I8E
 
 | | Acyl-ACP synthetase structure bound to C18:1-ACP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl carrier protein, Acyl-acyl carrier protein synthetase, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
7U15
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type B case 2 (case 7). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U16
 
 | | TMEM106B(120-254) protofilament from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (all cases combined). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U17
 
 | | TMEM106B(120-254) T185S singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type B case 2 (case 7). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U12
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (case 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U18
 
 | | TMEM106B(120-254) T185S protofilament from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (all cases combined). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TMEM106B protein | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-20 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U0Z
 
 | | High-resolution map of tau filament from progressive supranuclear palsy (PSP) case 1 | | Descriptor: | Isoform Tau-E of Microtubule-associated protein tau | | Authors: | FItzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U10
 
 | | TMEM106B(120-254) protofilament from progressive supranuclear palsy (PSP) case 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U14
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type C (case 8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7U11
 
 | | TMEM106B(120-254) protofilament from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type A (case 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
