5FE9
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1122 (compound 13) | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2B, ~{N}-(1,4-dimethyl-2-oxidanylidene-quinolin-7-yl)methanesulfonamide | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
3CYN
 
 | | The structure of human GPX8 | | Descriptor: | GLYCEROL, Probable glutathione peroxidase 8, SULFATE ION | | Authors: | Kavanagh, K.L, Johansson, C, Yue, W.W, Kochan, G, Pike, A.C.W, Murray, J, Roos, A.K, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GPX8
To be Published
|
|
5FDZ
 
 | | Crystal structure of human PCAF bromodomain in complex with compound BDOMB00091a (compound 14) | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2B, ~{N}-methyl-2-(oxan-4-yloxy)-5-(2-oxidanylidene-2-phenylazanyl-ethoxy)benzamide | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE3
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment MB360 (fragment 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1,2-benzoxazol-3-amine, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE5
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment MB093 (fragment 7) | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(1,2,3-thiadiazol-4-yl)phenyl]methanamine, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE4
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment MB364 (fragment 5) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1,4-benzodioxine-5-carboxamide, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
4AQL
 
 | | HUMAN GUANINE DEAMINASE IN COMPLEX WITH VALACYCLOVIR | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl L-valinate, GUANINE DEAMINASE, ZINC ION | | Authors: | Welin, M, Egeblad, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, Weigelt, J, Nordlund, P. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Pan-Pathway Based Interaction Profiling of Fda-Approved Nucleoside and Nucleobase Analogs with Enzymes of the Human Nucleotide Metabolism.
Plos One, 7, 2012
|
|
5FE1
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment BR004 (fragment 1) | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE8
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1126 (compound 12) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
3QMC
 
 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*CP*CP*GP*CP*TP*GP*GP*C)-3', 5'-D(*GP*CP*CP*AP*GP*CP*GP*GP*TP*GP*GP*C)-3', CpG-binding protein, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3QMB
 
 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3', CALCIUM ION, CpG-binding protein, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3CEY
 
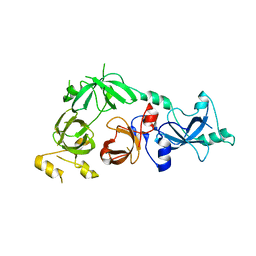 | | Crystal structure of L3MBTL2 | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein | | Authors: | Nady, N, Guo, Y, Pan, P, Allali-Hassani, A, Qi, C, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Bochkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
5IN3
 
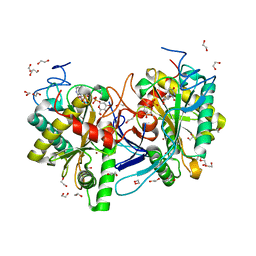 | | Crystal structure of glucose-1-phosphate bound nucleotidylated human galactose-1-phosphate uridylyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Kopec, J, McCorvie, T, Tallant, C, Velupillai, S, Shrestha, L, Fitzpatrick, F, Patel, D, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis of classic galactosemia from the structure of human galactose 1-phosphate uridylyltransferase.
Hum.Mol.Genet., 25, 2016
|
|
4JXT
 
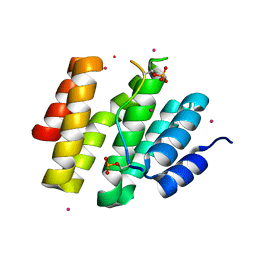 | | CID of human RPRD1A in complex with a phosphorylated peptide from RPB1-CTD | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Regulation of nuclear pre-mRNA domain-containing protein 1A, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5HTC
 
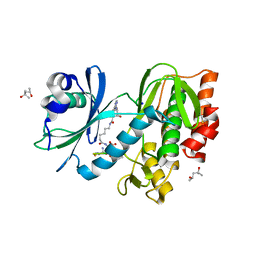 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3372 | | Descriptor: | (2R)-2-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}butanedioic acid (non-preferred name), (4S)-2-METHYL-2,4-PENTANEDIOL, ARC-3372 INHIBITOR, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3QMD
 
 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | CpG-binding protein, DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*AP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*TP*CP*GP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3F3S
 
 | | The Crystal Structure of Human Lambda-Crystallin, CRYL1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Lambda-crystallin homolog, ... | | Authors: | Ugochukwu, E, Johansson, C, Yue, W.W, Kochan, G, Pilka, E, Kramm, A, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Lambda-Crystallin, CRYL1
To be Published
|
|
3F70
 
 | | Crystal structure of L3MBTL2-H4K20me1 complex | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein, N-METHYL-LYSINE | | Authors: | Guo, Y, Qi, C, Allali-Hassani, A, Pan, P, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Botchkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
4AP2
 
 | | Crystal structure of the human KLHL11-Cul3 complex at 2.8A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CULLIN-3, ... | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Filippakopoulos, P, Ayinampudi, V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
5MYG
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with NI-57 chemical probe | | Descriptor: | 4-cyano-~{N}-(1,3-dimethyl-2-oxidanylidene-quinolin-6-yl)-2-methoxy-benzenesulfonamide, Peregrin | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Krojer, T, Nunez-Alonso, G, Kopec, J, Fitzpatrick, F, Savitsky, P, Fedorov, O, Brennan, P.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of a Chemical Probe for the Bromodomain and Plant Homeodomain Finger-Containing (BRPF) Family of Proteins.
J. Med. Chem., 60, 2017
|
|
4APF
 
 | | Crystal structure of the human KLHL11-Cul3 complex at 3.1A resolution | | Descriptor: | CULLIN 3, KELCH-LIKE PROTEIN 11 | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Vollmar, M, Ugochukwu, E, Muniz, J.R.C, Ayinampudi, V, Savitsky, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
6T28
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6T29
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
5FE0
 
 | | Crystal structure of human PCAF bromodomain in complex with acetyllysine | | Descriptor: | Histone acetyltransferase KAT2B, N(6)-ACETYLLYSINE | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE7
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment ZB2216 (fragment 11) | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-hydroxyethyl)-3-methyl-6,7-dihydro-5~{H}-indazol-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
