7KTQ
 
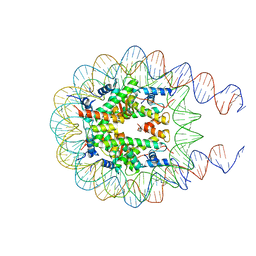 | | Nucleosome from a dimeric PRC2 bound to a nucleosome | | Descriptor: | 601 DNA (167-MER), Histone H2A, Histone H2B, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
5YGK
 
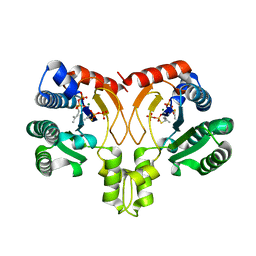 | | Crystal structure of a synthase from Streptomyces sp. CL190 with dmaspp | | Descriptor: | Cyclolavandulyl diphosphate synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-09-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Catalytic Role of Conserved Asparagine, Glutamine, Serine, and Tyrosine Residues in Isoprenoid Biosynthesis Enzymes.
Acs Catalysis, 8, 2018
|
|
5Y1Y
 
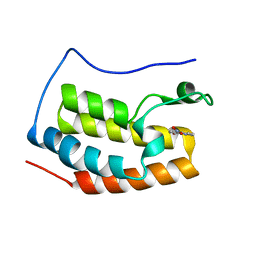 | |
5YRN
 
 | | Structure of RIP2 CARD domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Wu, B, Gong, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RIP2 activation and signaling.
Nat Commun, 9, 2018
|
|
5CF4
 
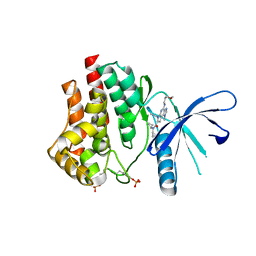 | | CRYSTAL STRUCTURE OF JANUS KINASE 2 IN COMPLEX WITH N,N-DICYCLOPROPYL-10-ETHYL-7-[(3-METHOXYPROPYL)AMINO] -3-METHYL-3,5,8,10-TETRAAZATRICYCLO[7.3.0.0,6] DODECA-1(9),2(6),4,7,11-PENTAENE-11-CARBOXAMIDE | | Descriptor: | N,N-dicyclopropyl-6-ethyl-4-[(3-methoxypropyl)amino]-1-methyl-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5YGJ
 
 | | Crystal structure of a synthase from Streptomyces sp. CL190 | | Descriptor: | Cyclolavandulyl diphosphate synthase | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-09-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Catalytic Role of Conserved Asparagine, Glutamine, Serine, and Tyrosine Residues in Isoprenoid Biosynthesis Enzymes.
Acs Catalysis, 8, 2018
|
|
5CF6
 
 | | CRYSTAL STRUCTURE OF JANUS KINASE 2 IN COMPLEX WITH N,N-DICYCLOPROPYL-10-[(2S)-2,3-DIHYDROXYPROPYL]-3-METHYL-7-(METHYLAMINO)-3,5,8,10-TETRAAZATRICYCLO [7.3.0.02,6]DODECA-1(9),2(6),4,7,11-PENTAENE-11-CARBOXAMIDE | | Descriptor: | N,N-dicyclopropyl-6-[(2S)-2,3-dihydroxypropyl]-1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5H21
 
 | | Trimethoxy-ring inhibitor in complex with the first bromodomain of BRD4 | | Descriptor: | 3,4,5-trimethoxy-~{N}-(2-thiophen-2-ylethyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of novel trimethoxy-ring BRD4 bromodomain inhibitors: AlphaScreen assay, crystallography and cell-based assay.
Medchemcomm, 8, 2017
|
|
3IFT
 
 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
5CZJ
 
 | |
7C01
 
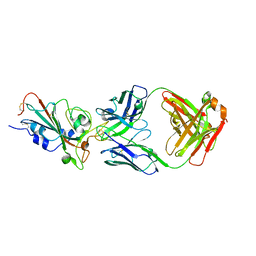 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
5Z4Y
 
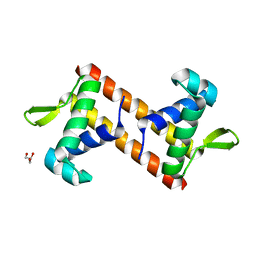 | |
7BSX
 
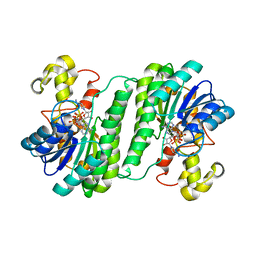 | | SDR protein NapW-NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Wen, W.H, Tang, G.L. | | Deposit date: | 2020-03-31 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.
Nat Commun, 12, 2021
|
|
7BTM
 
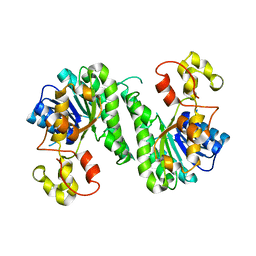 | | SDR protein/resistance protein NapW | | Descriptor: | Short chain dehydrogenase | | Authors: | Wen, W.H, Tang, G.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08311653 Å) | | Cite: | Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.
Nat Commun, 12, 2021
|
|
5DJO
 
 | | Crystal structure of the CC1-FHA tandem of Kinesin-3 KIF13A | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Ren, J.Q, Li, W, Huo, L, Feng, W. | | Deposit date: | 2015-09-02 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Correlation of the Neck Coil with the Coiled-coil (CC1)-Forkhead-associated (FHA) Tandem for Active Kinesin-3 KIF13A
J.Biol.Chem., 291, 2016
|
|
3K9U
 
 | | Crystal structure of paia acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
5Z50
 
 | | Crystal structure of PaCysB regulatory domain | | Descriptor: | Cys regulon transcriptional activator, GLYCEROL, SULFATE ION | | Authors: | Yang, C, Liang, H, Gan, J. | | Deposit date: | 2018-01-15 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa.
Mol.Microbiol., 111, 2019
|
|
5Z4Z
 
 | |
5XK9
 
 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with GSPP and DMAPP | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8D36
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
3HZG
 
 | | Crystal structure of mycobacterium tuberculosis thymidylate synthase X bound with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Staker, B.L, Rathod, P, Hunter, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increasing the structural coverage of tuberculosis drug targets.
Tuberculosis (Edinb), 95, 2015
|
|
3HWI
 
 | |
7KSO
 
 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
