6F3X
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide in frozen DDM/CHS detergent micelle solution determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3Y
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide bound to human Bradykinin 1 Receptor (B1R) determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
4O9H
 
 | | Structure of Interleukin-6 in complex with a Camelid Fab fragment | | Descriptor: | Heavy Chain of the Camelid Fab fragment 61H7, Interleukin-6, Light Chain of the Camelid Fab fragment 61H7 | | Authors: | Klarenbeek, A, Blanchetot, C, Schragel, G, Sadi, A.S, Ongenae, N, Hemrika, W, Wijdenes, J, Spinelli, S, Desmyter, A, Cambillau, C, Hultberg, A, Kretz-rommel, A, Dreier, T, De haard, H.J.W, Roovers, R.C. | | Deposit date: | 2014-01-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Combining residues of naturally-occurring Camelid somatic affinity variants yields ultra-potent human therapeutic IL-6 antibodies
To be Published
|
|
2X8K
 
 | | Crystal Structure of SPP1 Dit (gp 19.1) Protein, a Paradigm of Hub Adsorption Apparatus in Gram-positive Infecting Phages. | | Descriptor: | HYPOTHETICAL PROTEIN 19.1 | | Authors: | Veesler, D, Robin, G, Lichiere, J, Auzat, I, Tavares, P, Bron, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Bacteriophage Spp1 Distal Tail Protein (Gp 19.1): A Baseplate Hub Paradigm in Gram+ Infecting Phages.
J.Biol.Chem., 285, 2010
|
|
2XF5
 
 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
1I3U
 
 | | THREE-DIMENSIONAL STRUCTURE OF A LLAMA VHH DOMAIN COMPLEXED WITH THE DYE RR1 | | Descriptor: | 5-(4,6-DIAMINO-[1,3,5]TRIAZIN-2-YLAMINO)-4-HYDROXY-3-(2-SULFO-PHENYLAZO)-NAPHTHALENE-2,7-DISULFONIC ACID, ANTIBODY VHH LAMA DOMAIN, SULFATE ION | | Authors: | Spinelli, S, Tegoni, M, Frenken, L, van Vliet, C, Cambillau, C. | | Deposit date: | 2001-02-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lateral recognition of a dye hapten by a llama VHH domain.
J.Mol.Biol., 311, 2001
|
|
1I3V
 
 | | THREE-DIMENSIONAL STRUCTURE OF A LAMA VHH DOMAIN UNLIGANDED | | Descriptor: | ANTIBODY VHH LAMA DOMAIN | | Authors: | Spinelli, S, Tegoni, M, Frenken, L, van Vliet, C, Cambillau, C. | | Deposit date: | 2001-02-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lateral recognition of a dye hapten by a llama VHH domain.
J.Mol.Biol., 311, 2001
|
|
4QGY
 
 | | Camelid (llama) nanobody n25 (VHH) against type 6 secretion system TssM protein | | Descriptor: | nanobody n25, VH domain | | Authors: | Nguyen, V.S, Desmyter, A, Le, T.T.H, Durand, E, Kellenberger, C, Douzi, B, Spinelli, S, Cascales, E, Cambillau, C, Roussel, A. | | Deposit date: | 2014-05-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody.
Plos One, 10, 2015
|
|
4QLR
 
 | | Llama nanobody n02 raised against EAEC T6SS TssM | | Descriptor: | Llama nanobody n02 VH domain | | Authors: | Nguyen, V.S, Desmyter, A, Le, T.T.H, Durand, E, Kellenberger, C, Douzi, B, Spinelli, S, Cascales, E, Cambillau, C, Roussel, A. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody.
Plos One, 10, 2015
|
|
3BFH
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with hexadecanoic acid | | Descriptor: | CHLORIDE ION, PALMITIC ACID, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
4R90
 
 | | Anti CD70 Llama glama Fab 27B3 | | Descriptor: | Anti CD70 Llama glama Fab 27B3 Heavy chain, Anti CD70 Llama glama Fab 27B3 Light chain, CALCIUM ION, ... | | Authors: | Klarenbeek, A, El Mazouari, K, Desmyter, A, Blanchetot, C, Hultberg, A, Roovers, R.C, Cambillau, C, Spinelli, S, Del-Favero, J, Verrips, T, de Haard, H, Achour, I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform.
MAbs, 7, 2015
|
|
4R96
 
 | | Structure of a Llama Glama Fab 48A2 against human cMet | | Descriptor: | Llama glama Fab 48A2 against human cMet H chain, Llama glama Fab 48A2 against human cMet L chain | | Authors: | Klarenbeek, A, El Mazouari, K, Desmyter, A, Blanchetot, C, Hultberg, A, Roovers, R.C, Cambillau, C, Spinelli, S, Del-Favero, J, Verrips, T, de Haard, H, Achour, I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform.
MAbs, 7, 2015
|
|
3BFB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the 9-keto-2(E)-decenoic acid | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
1QD0
 
 | | CAMELID HEAVY CHAIN VARIABLE DOMAINS PROVIDE EFFICIENT COMBINING SITES TO HAPTENS | | Descriptor: | 3-HYDROXY-7-(4-{1-[2-HYDROXY-3-(2-HYDROXY-5-SULFO-PHENYLAZO)-BENZYL]-2-SULFO-ETHYLAMINO}-[1,2,5]TRIAZIN-2-YLAMINO)-2-(2-HYDROXY-5-SULFO-PHENYLAZO)-NAPTHALENE-1,8-DISULFONIC ACID, COPPER (II) ION, VHH-R2 ANTI-RR6 ANTIBODY | | Authors: | Spinelli, S, Frenken, L.G.J, Hermans, P, Verrips, T, Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 1999-07-08 | | Release date: | 2000-07-19 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Camelid heavy-chain variable domains provide efficient combining sites to haptens.
Biochemistry, 39, 2000
|
|
1P4I
 
 | | Crystal Structure of scFv against peptide GCN4 | | Descriptor: | ANTIBODY VARIABLE LIGHT CHAIN, antibody variable heavy chain | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1WND
 
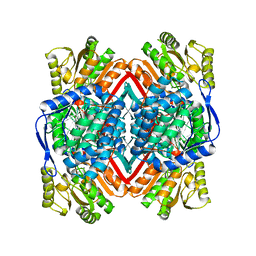 | | Escherichia coli YdcW gene product is a medium-chain aldehyde dehydrogenase as determined by kinetics and crystal structure | | Descriptor: | CALCIUM ION, Putative betaine aldehyde dehydrogenase | | Authors: | Gruez, A, Roig-Zamboni, V, Tegoni, M, Cambillau, C. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Kinetics Identify Escherichia coli YdcW Gene Product as a Medium-chain Aldehyde Dehydrogenase
J.Mol.Biol., 343, 2004
|
|
3EJC
 
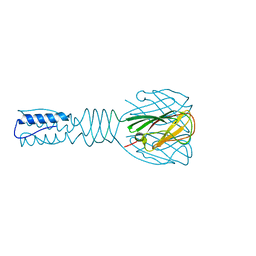 | | Full length Receptor Binding Protein from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein (BPP) | | Authors: | Spinelli, S, Lichiere, J, Blangy, S, Sciara, G, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and molecular assignment of lactococcal phage TP901-1 baseplate.
J.Biol.Chem., 285, 2010
|
|
1WNB
 
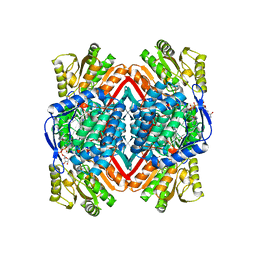 | | Escherichia coli YdcW gene product is a medium-chain aldehyde dehydrogenase (complexed with nadh and betaine aldehyde) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETAINE ALDEHYDE, Putative betaine aldehyde dehydrogenase | | Authors: | Gruez, A, Roig-Zamboni, V, Tegoni, M, Cambillau, C. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure and Kinetics Identify Escherichia coli YdcW Gene Product as a Medium-chain Aldehyde Dehydrogenase
J.Mol.Biol., 343, 2004
|
|
2DJF
 
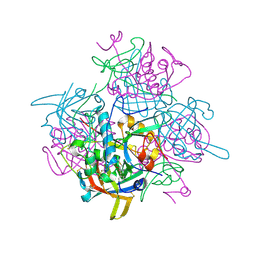 | | Crystal Structure of human dipeptidyl peptidase I (Cathepsin C) in complex with the inhibitor Gly-Phe-CHN2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Molgaard, A, Arnau, J, Lauritzen, C, Larsen, S, Petersen, G, Pedersen, J. | | Deposit date: | 2006-04-02 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2
Biochem.J., 401, 2007
|
|
2DJG
 
 | | Re-determination of the native structure of human dipeptidyl peptidase I (cathepsin C) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Dipeptidyl-peptidase 1, ... | | Authors: | Molgaard, A, Arnau, J, Lauritzen, C, Larsen, S, Petersen, G, Pedersen, J. | | Deposit date: | 2006-04-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2
Biochem.J., 401, 2007
|
|
6QDW
 
 | | Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Hodirnau, V.V, Kudlinzki, D, Mao, J, Glaubitz, C, Frangakis, A, Schwalbe, H. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cysteine oxidation and disulfide formation in the ribosomal exit tunnel.
Nat Commun, 11, 2020
|
|
2BSD
 
 | | Structure of Lactococcal Bacteriophage p2 Receptor Binding Protein | | Descriptor: | RECEPTOR BINDING PROTEIN | | Authors: | Spinelli, S, Desmyter, A, Verrips, C.T, Dehaard, H.J.W, Moineau, S, Cambillau, C. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lactococcal Bacteriophage P2 Receptor-Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1P4B
 
 | | Three-Dimensional Structure Of a Single Chain Fv Fragment Complexed With The peptide GCN4(7P-14P). | | Descriptor: | Antibody Variable heavy chain, Antibody Variable light chain, GCN4(7P-14P) peptide | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-22 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1PT7
 
 | | Crystal structure of the apo-form of the yfdW gene product of E. coli | | Descriptor: | GLYCEROL, Hypothetical protein yfdW, PHOSPHATE ION | | Authors: | Gruez, A, Roig-Zamboni, V, Valencia, C, Campanacci, V, Cambillau, C. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the Escherichia coli yfdW gene product reveals a New fold of two interlaced rings identifying a wide family of CoA transferases.
J.Biol.Chem., 278, 2003
|
|
3FBL
 
 | | Crystal structure of ORF132 of the archaeal virus Acidianus Filamentous Virus 1 (AFV1) | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein | | Authors: | Goulet, A, Leulliot, N, Prangishvili, D, van Tilbeurgh, H, Campanacci, V, Cambillau, C. | | Deposit date: | 2008-11-19 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Acidianus filamentous virus 1 coat proteins display a helical fold spanning the filamentous archaeal viruses lineage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
