2XMU
 
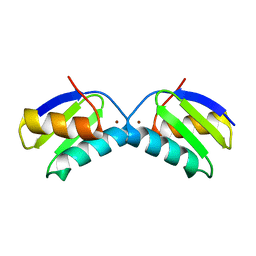 | | Copper chaperone Atx1 from Synechocystis PCC6803 (Cu2 form) | | Descriptor: | COPPER (I) ION, SSR2857 PROTEIN | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
6ZLN
 
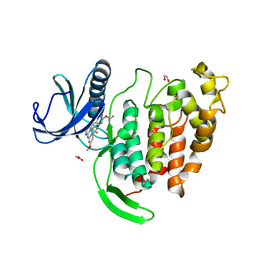 | | CLK1 bound with GW807982X (Cpd 8) | | Descriptor: | 1,2-ETHANEDIOL, 4-(6-ethoxypyrazolo[1,5-b]pyridazin-3-yl)-~{N}-[3-methoxy-5-(trifluoromethyl)phenyl]pyrimidin-2-amine, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6ZLA
 
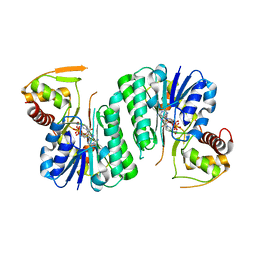 | |
2XN3
 
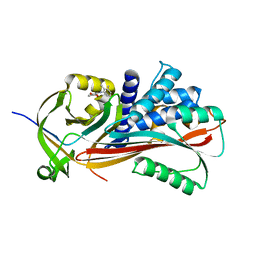 | | Crystal structure of thyroxine-binding globulin complexed with mefenamic acid | | Descriptor: | 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, THYROXINE-BINDING GLOBULIN | | Authors: | Qi, X, Yan, Y, Wei, Z, Zhou, A. | | Deposit date: | 2010-07-30 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric Modulation of Hormone Release from Thyroxine and Corticosteroid Binding-Globulins.
J.Biol.Chem., 286, 2011
|
|
2XQB
 
 | | Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 | | Descriptor: | ANTI-IL-15 ANTIBODY, INTERLEUKIN 15, SULFATE ION | | Authors: | Lowe, D.C, Gerhardt, S, Ward, A, Hargreaves, D, Anderson, M, StGallay, S, Vousden, K, Ferraro, F, Pauptit, R.A, Cochrane, D, Pattison, D.V, Buchanan, C, Popovic, B, Finch, D.K, Wilkinson, T, Sleeman, M, Vaughan, T.J, Cruwys, S, Mallinder, P.R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
J.Mol.Biol., 406, 2011
|
|
2XYT
 
 | | Crystal structure of Aplysia californica AChBP in complex with d- tubocurarine | | Descriptor: | D-TUBOCURARINE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
6ZD8
 
 | | Crystal structure of YTHDC1 T379V mutant | | Descriptor: | SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
2Y31
 
 | | Simocyclinone C4 bound form of TetR-like repressor SimR | | Descriptor: | CALCIUM ION, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
6ZH9
 
 | | Ternary complex CR3022 H11-H4 and RBD (SARS-CoV-2) | | Descriptor: | CR3022 Light chain, CR3022 heavy, Nanobody H11-H4, ... | | Authors: | Naismith, J.H, Mikolajek, H, Le Bas, A. | | Deposit date: | 2020-06-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZIH
 
 | |
6ZLK
 
 | | Equilibrium Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid/UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
2XLS
 
 | | Joint-functions of protein residues and NADP(H) in oxygen-activation by flavin-containing monooxygenase: Asn78Lys mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Orru, R, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Joint-Functions of Protein Residues and Nadp(H) in Oxygen-Activation by Flavin-Containing Monooxygenase
J.Biol.Chem., 285, 2010
|
|
2XMM
 
 | | Visualising the Metal-binding Versatility of Copper Trafficking Sites: H61Y Atx1 side-to-side | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SODIUM ION, ... | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
2XXA
 
 | | The Crystal Structure of the Signal Recognition Particle (SRP) in Complex with its Receptor(SR) | | Descriptor: | 4.5S RNA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ataide, S.F, Schmitz, N, Shen, K, Ke, A, Shan, S, Doudna, J.A, Ban, N. | | Deposit date: | 2010-11-09 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | The Crystal Structure of the Signal Recognition Particle in Complex with its Receptor.
Science, 331, 2011
|
|
2XP4
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 2-phenyl-1H-imidazole-4-carboxylic acid, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2Y00
 
 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND PARTIAL AGONIST DOBUTAMINE (CRYSTAL DOB92) | | Descriptor: | BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, DOBUTAMINE, ... | | Authors: | Warne, A, Moukhametzianov, R, Baker, J.G, Nehme, R, Edwards, P.C, Leslie, A.G.W, Schertler, G.F.X, Tate, C.G. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor
Nature, 469, 2011
|
|
6ZIW
 
 | | The IRAK3 Pseudokinase Domain Bound To ATPgammaS | | Descriptor: | Interleukin-1 receptor-associated kinase 3, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Kraemer, A, Knapp, S. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The IRAK3 Pseudokinase Domain Bound To ATPgammaS
To Be Published
|
|
6ZJJ
 
 | |
2Y0J
 
 | | Triazoloquinazolines as a novel class of phosphodiesterase 10A (PDE10A) inhibitors, part 2, Lead-optimisation. | | Descriptor: | 5-(1H-BENZIMIDAZOL-2-YLMETHYLSULFANYL)-2-METHYL-[1,2,4]TRIAZOLO[1,5-C]QUINAZOLINE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Kehler, J, Ritzen, A, Langgard, M, Petersen, S.L, Christoffersen, C.T, Nielsen, J, Kilburn, J.P. | | Deposit date: | 2010-12-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Triazoloquinazolines as a Novel Class of Phosphodiesterase 10A (Pde10A) Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6ZLH
 
 | | the structure of glutamate transporter homologue GltTk in complex with the photo switchable compound (trans) | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[4-[2-(4-methoxyphenyl)hydrazinyl]phenyl]methoxy]butanedioic acid, DECYL-BETA-D-MALTOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arkhipova, V, Slotboom, D.J, Guskov, A. | | Deposit date: | 2020-06-30 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Aspects of Photopharmacology: Insight into the Binding of Photoswitchable and Photocaged Inhibitors to the Glutamate Transporter Homologue.
J.Am.Chem.Soc., 143, 2021
|
|
2XW6
 
 | | The Crystal Structure of Methylglyoxal Synthase from Thermus sp. GH5 Bound to Phosphate Ion. | | Descriptor: | METHYLGLYOXAL SYNTHASE, PHOSPHATE ION | | Authors: | Shahsavar, A, Erfani Moghaddam, M, Antonyuk, S.V, Khajeh, K, Naderi-Manesh, H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Atomic Resolution Structure of Methylglyoxal Synthase from Thermus Sp. Gh5 Bound to Phosphate: Insights Into the Distinctive Effects of Phosphate on the Enzyme Structure
To be Published
|
|
2XYE
 
 | | HIV-1 Inhibitors with a Tertiary-Alcohol-containing Transition-State Mimic and various P2 and P1 prime Substituents | | Descriptor: | METHYL N-[(2S)-1-[2-[(4R)-5-[[(2S)-3,3-DIMETHYL-1-METHYLAMINO-1-OXO-BUTAN-2-YL]AMINO]-4-HYDROXY-5-OXO-4-(PHENYLMETHYL)PENTYL]-2-[(4-PHENYLPHENYL)METHYL]HYDRAZINYL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, PROTEASE | | Authors: | Ohrngren, P, Wu, X, Persson, M, Ekegren, J.K, Wallberg, H, Rosenquist, A, Samuelsson, B, Unge, T, Larhed, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors with a Tertiary Alcohol Containing Transition-State Mimic and Various P2 and P1' Substituents
Med.Chem.Commun., 2, 2011
|
|
2Y05
 
 | | Crystal structure of human leukotriene B4 12-hydroxydehydrogenase in complex with NADP and raloxifene | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yue, W.W, Shafqat, N, Krojer, T, Pike, A.C.W, von Delft, F, Sethi, R, Savitsky, P, Johansson, C, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Human Leukotriene B4 12-Hydroxydehydrogenase in Complex with Nadp and Raloxifene
To be Published
|
|
