5T0U
 
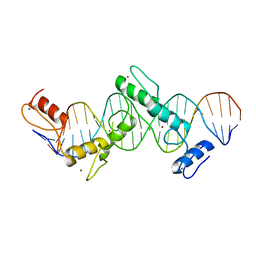 | | CTCF ZnF2-7 and DNA complex structure | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
3NFC
 
 | |
5CN0
 
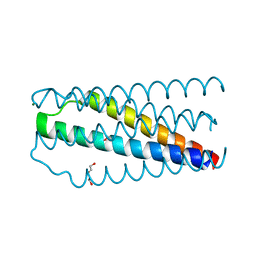 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5TJ8
 
 | |
5CMU
 
 | |
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
5TJ7
 
 | |
5TJQ
 
 | | Structure of WWP2 2,3-linker-HECT | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2,NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2016-10-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Tunable Brake for HECT Ubiquitin Ligases.
Mol. Cell, 66, 2017
|
|
6MBJ
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, P21 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6LKO
 
 | |
6WI7
 
 | | RING1B-BMI1 fusion in closed conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1 chimera, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
6WI8
 
 | |
6LYG
 
 | |
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6MG1
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta, ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MBK
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, First P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MBL
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, Second P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MG3
 
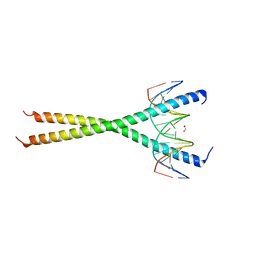 | | V285A Mutant of the C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MG2
 
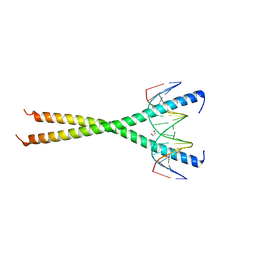 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2221 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
7SFF
 
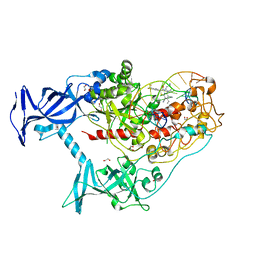 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3852279B | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]sulfanyl}-2-[4-(trifluoromethyl)phenyl]acetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFC
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3735967A | | Descriptor: | 1,2-ETHANEDIOL, DNA (12-MER), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
6LWA
 
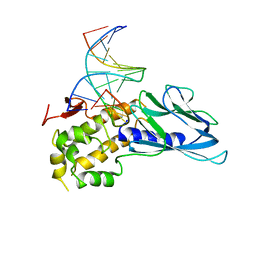 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
7SFD
 
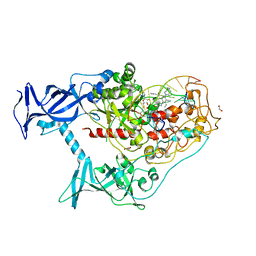 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3543105A | | Descriptor: | (2S)-2-({3,5-dicyano-4-ethyl-6-[4-(2-hydroxyethyl)-1,4-diazepan-1-yl]pyridin-2-yl}sulfanyl)-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
6LWF
 
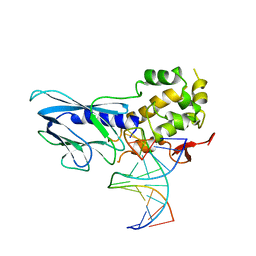 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing guanidinohydantoin (Gh) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
