7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
3JTO
 
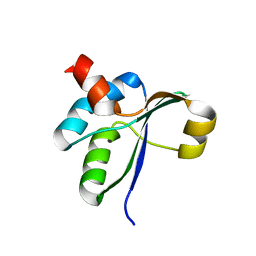 | | Crystal structure of the c-terminal domain of YpbH | | Descriptor: | Adapter protein mecA 2 | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the MecA Degradation Tag
To be Published
|
|
2MOX
 
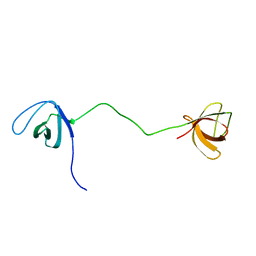 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
3P12
 
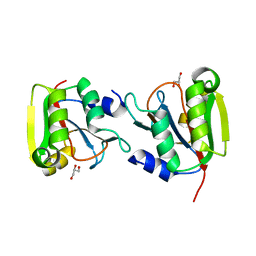 | | Crystal Structure of D-ribose Pyranase Sa240 | | Descriptor: | D-ribose pyranase, GLYCEROL | | Authors: | Wu, M, Wang, L, Zang, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Sa240: A ribose pyranase homolog with partial active site from Staphylococcus aureus
J.Struct.Biol., 174, 2011
|
|
5Y4M
 
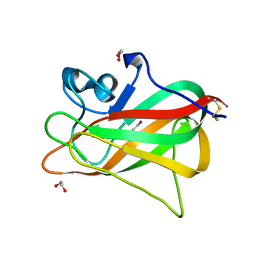 | | Discoidin domain of human CASPR2 | | Descriptor: | 1,2-ETHANEDIOL, human CASPR2 Disc domain | | Authors: | Liu, H, Xu, F, Zhang, J, Liang, W. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural mapping of hot spots within human CASPR2 discoidin domain for autoantibody recognition.
J. Autoimmun., 96, 2019
|
|
5YEG
 
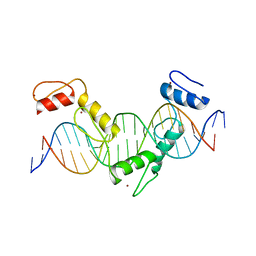 | | Crystal structure of CTCF ZFs4-8-Hs5-1a complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*AP*AP*CP*CP*AP*GP*CP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*GP*CP*TP*GP*GP*TP*TP*AP*AP*AP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
3P13
 
 | |
7DR8
 
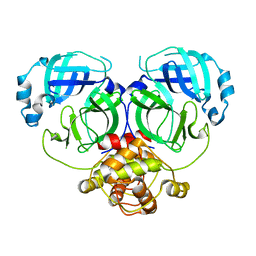 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
1XRG
 
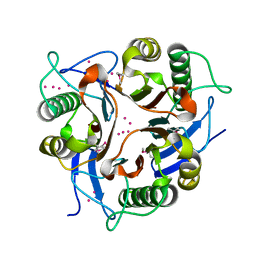 | | Conserved hypothetical protein from Clostridium thermocellum Cth-2968 | | Descriptor: | Putative translation initiation inhibitor, yjgF family, UNKNOWN ATOM OR ION | | Authors: | Zhao, M, Chang, J, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Zhang, H, Arendall III, W.B, Ljundahl, L, Liu, Z.-J, Rose, J, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-14 | | Release date: | 2004-12-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved hypothetical protein from Clostridium thermocellum Cth-2968
To be published
|
|
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural insights reveal interplay between LAG-3 homodimerization, ligand binding, and function.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3JTN
 
 | | Crystal Structure of the c-terminal domain of YpbH | | Descriptor: | Adapter protein mecA 2, IODIDE ION | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the MecA degradation tag
To be Published
|
|
3JTP
 
 | | crystal structure of the C-terminal domain of MecA | | Descriptor: | Adapter protein mecA 1, IODIDE ION | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | crystal structure of the MecA degradation tag
To be Published
|
|
4EAJ
 
 | | Co-crystal of AMPK core with AMP soaked with ATP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
8IO0
 
 | | Cryo-EM structure of human HCN3 channel with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure of human HCN3 channel and its regulation by cAMP.
J.Biol.Chem., 300, 2024
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
4R7L
 
 | | Structure of Human Leukotriene A4 Hydrolase in complex with inhibitor H1 | | Descriptor: | ACETATE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Ouyang, P, Cui, K, Lu, W, Huang, J. | | Deposit date: | 2014-08-27 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Human Leukotriene A4 Hydrolase in complex with SAHA
To be Published
|
|
7N15
 
 | | Structure of TAX-4_R421W w/cGMP open state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N16
 
 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N17
 
 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
2PPH
 
 | | solution structure of human MEKK3 PB1 domain | | Descriptor: | Mitogen-activated protein kinase kinase kinase 3 | | Authors: | Hu, Q, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insight into the Binding Properties of MEKK3 PB1 to MEK5 PB1 from Its Solution Structure.
Biochemistry, 46, 2007
|
|
7DVM
 
 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
5YEL
 
 | | Crystal structure of CTCF ZFs6-11-gb7CSE | | Descriptor: | DNA (26-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
1ZGH
 
 | | Methionyl-tRNA formyltransferase from Clostridium thermocellum | | Descriptor: | Methionyl-tRNA formyltransferase, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Kataeva, I, Xu, H, Zhao, M, Chang, J, Liu, Z, Chen, L, Tempel, W, Habel, J, Zhou, W, Lee, D, Lin, D, Chang, S, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionyl-tRNA formyltransferase from Clostridium thermocellum
To be published
|
|
