6O9C
 
 | |
3E37
 
 | |
3E27
 
 | |
3E32
 
 | | Protein farnesyltransferase complexed with FPP and ethylenediamine scaffold inhibitor 2 | | Descriptor: | FARNESYL DIPHOSPHATE, N-benzyl-N-(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)-1-methyl-1H-imidazole-4-sulfonamide, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
3E34
 
 | | Protein farnesyltransferase complexed with FPP and ethylenediamine-scaffold inhibitor 10 | | Descriptor: | 3-{2'-[{[1-(tert-butoxycarbonyl)piperidin-4-yl]methyl}(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)sulfamoyl]biphenyl-3-yl}propanoic acid, FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
3E30
 
 | | Protein farnesyltransferase complexed with FPP and ethylene diamine inhibitor 4 | | Descriptor: | FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
3F4C
 
 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10, with glycerol bound | | Descriptor: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
3FZO
 
 | | Crystal Structure of PYK2-Apo, Proline-rich Tyrosine Kinase | | Descriptor: | Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZR
 
 | | Crystal structure of PYK2 complexed with PF-431396 | | Descriptor: | N-methyl-N-{2-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]phenyl}methanesulfonamide, PHOSPHATE ION, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZP
 
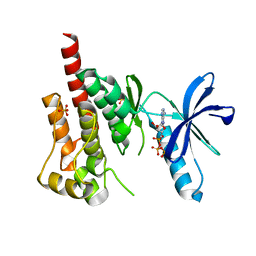 | | Crystal structure of PYK2 complexed with ATPgS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein tyrosine kinase 2 beta, SULFATE ION | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZS
 
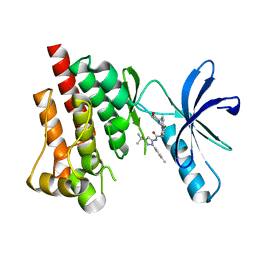 | | Crystal Structure of PYK2 complexed with BIRB796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZT
 
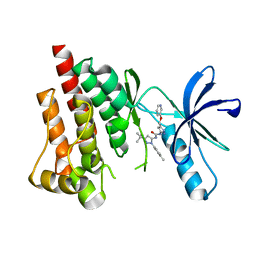 | | Crystal structure of PYK2 complexed with PF-4618433 | | Descriptor: | 1-[5-tert-butyl-2-(4-methylphenyl)-1,2-dihydro-3H-pyrazol-3-ylidene]-3-{3-[(pyridin-3-yloxy)methyl]-1H-pyrazol-5-yl}urea, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3GE3
 
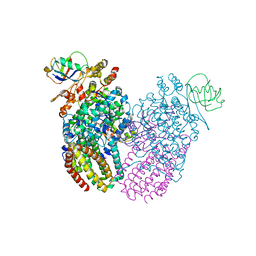 | | Crystal Structure of the reduced Toluene 4-Monooxygenase HD T201A mutant complex | | Descriptor: | ACETATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Elsen, N.L, Bailey, L.J, Fox, B.G. | | Deposit date: | 2009-02-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Role for threonine 201 in the catalytic cycle of the soluble diiron hydroxylase toluene 4-monooxygenase.
Biochemistry, 48, 2009
|
|
3GAB
 
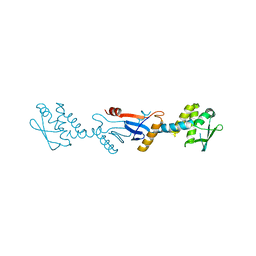 | | C-terminal domain of Bacillus subtilis MutL crystal form I | | Descriptor: | DNA mismatch repair protein mutL | | Authors: | Guarne, A, Pillon, M.C, Lorenowicz, J.J, Mitchell, R.R, Chung, Y.S, Friedhoff, P. | | Deposit date: | 2009-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the endonuclease domain of MutL: unlicensed to cut.
Mol.Cell, 39, 2010
|
|
3E33
 
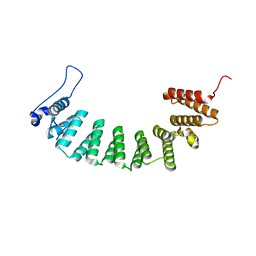 | | Protein farnesyltransferase complexed with FPP and ethylenediamine scaffold inhibitor 7 | | Descriptor: | FARNESYL DIPHOSPHATE, N-benzyl-N-(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)-2-methylbenzenesulfonamide, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
3DLK
 
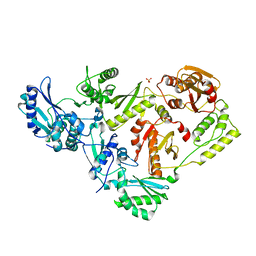 | | Crystal Structure of an engineered form of the HIV-1 Reverse Transcriptase, RT69A | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, p51 RT | | Authors: | Ho, W.C, Bauman, J.D, Himmel, D.M, Das, K, Arnold, E. | | Deposit date: | 2008-06-27 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design.
Nucleic Acids Res., 36, 2008
|
|
3H8V
 
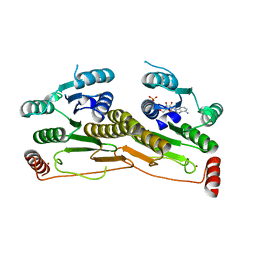 | | Human Ubiquitin-activating Enzyme 5 in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ubiquitin-like modifier-activating enzyme 5, ZINC ION | | Authors: | Walker, J.R, Bacik, J.P, Rastgoo, N, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: mechanistic insights into a minimalistic E1 enzyme.
J.Biol.Chem., 285, 2010
|
|
3GU1
 
 | | Y97W mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GTH
 
 | | D71G/E101G/M234I mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, FORMIC ACID, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GU2
 
 | | Y97L/G100-/E101- mutant in organophosphorus hydrolase | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GU9
 
 | | R228A mutation in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GX2
 
 | |
3GX6
 
 | |
3F4D
 
 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10 | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
3GK1
 
 | | X-ray structure of bovine SBi132,Ca(2+)-S100B | | Descriptor: | 2-[(5-hex-1-yn-1-ylfuran-2-yl)carbonyl]-N-methylhydrazinecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
