5DZN
 
 | | human T-cell immunoglobulin and mucin domain protein 4 | | Descriptor: | T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Yuan, S, Rao, Z, Wang, X. | | Deposit date: | 2015-09-25 | | Release date: | 2015-11-25 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope.
Protein Cell, 6, 2015
|
|
5DZO
 
 | |
1F6W
 
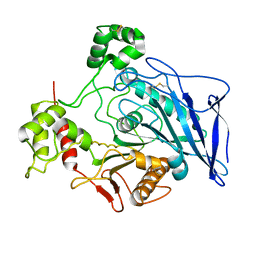 | |
1XMZ
 
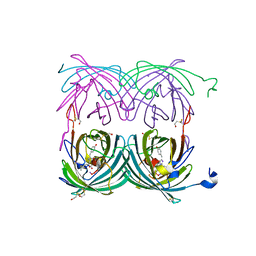 | | Crystal structure of the dark state of kindling fluorescent protein kfp from anemonia sulcata | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Quillin, M.L, Anstrom, D.M, Shu, X, O'Leary, S, Kallio, K, Chudakov, D.M, Remington, S.J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-04-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kindling Fluorescent Protein from Anemonia sulcata: Dark-State Structure at 1.38 Resolution
Biochemistry, 44, 2005
|
|
2MNY
 
 | | NMR Structure of KDM5B PHD1 finger | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
4EMP
 
 | | Crystal structure of the mutant of ClpP E137A from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Ye, F, Zhang, J, Liu, H, Luo, C, Yang, C.-G. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
J.Biol.Chem., 288, 2013
|
|
4EMM
 
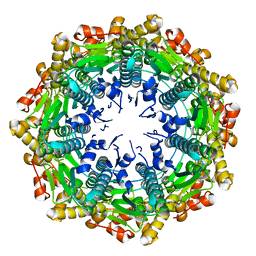 | |
7A3O
 
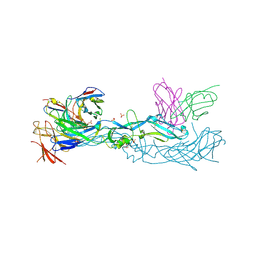 | | Crystal structure of dengue 1 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | Core protein, GLYCEROL, SULFATE ION, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3R
 
 | | Crystal structure of dengue 1 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3N
 
 | | Crystal structure of Zika virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | CALCIUM ION, Core protein, EDE1 C10 Fab | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3T
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | Core protein, EDE1 C8 antibody Fab fragment, GLYCEROL, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3U
 
 | | Crystal structure of Zika virus envelope glycoprotein in complex with the divalent F(ab')2 fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | EDE1 C10 antibody divalent F(ab')2 fragment, EDE1 C10 divalent F(ab')2 fragment, Envelope protein | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3P
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Single Chain Variable Fragment | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3S
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, SULFATE ION | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Navarro-Sanchez, E, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3Q
 
 | | Crystal structure of dengue 4 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
4R28
 
 | |
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
6UIP
 
 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
7BST
 
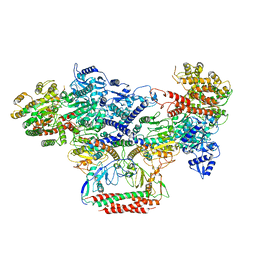 | | EcoR124I-Ocr in the Intermediate State | | Descriptor: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-03-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTR
 
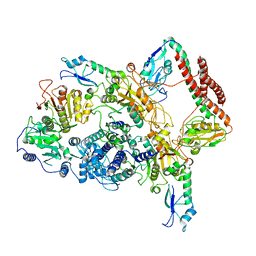 | | EcoR124I-ArdA in the Restriction-Alleviation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTO
 
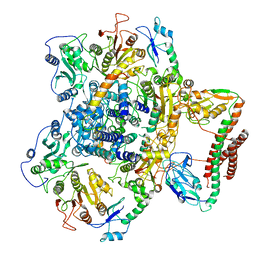 | | EcoR124I-ArdA in the Translocation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTQ
 
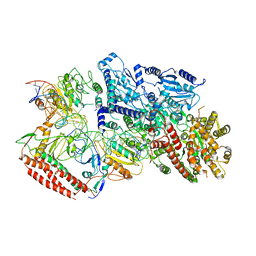 | | EcoR124I-DNA in the Restriction-Alleviation State | | Descriptor: | DNA (64-MER), Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
