7C8F
 
 | | Structure of alginate lyase AlyC3 in complex with dimannuronate(2M) | | Descriptor: | H127A/Y244A mutant of alginate lyase AlyC3 in complex with dimannuronate, MALONATE ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Zhang, Y.Z, Xu, F, Chen, X.L, Wang, P. | | Deposit date: | 2020-05-30 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic.
J.Biol.Chem., 295, 2020
|
|
6UMW
 
 | | Crystal structure of hEphB1 bound with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2019-10-10 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N5H
 
 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
7S4G
 
 | | Fab fragment bound to the Cter peptide of Ly6G6D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Lymphocyte antigen 6 complex locus protein G6d, ... | | Authors: | Rouge, L, Lupardus, P. | | Deposit date: | 2021-09-08 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Anti-LY6G6D/CD3 T-Cell-Dependent Bispecific Antibody for the Treatment of Colorectal Cancer.
Mol.Cancer Ther., 21, 2022
|
|
7M8K
 
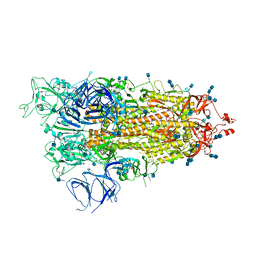 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
7VP9
 
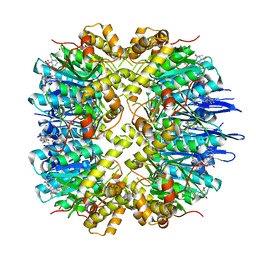 | | Crystal structure of human ClpP in complex with ZG111 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma.
Cell Chem Biol, 29, 2022
|
|
1FHR
 
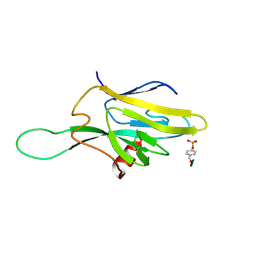 | | SOLUTION STRUCTURE OF THE FHA2 DOMAIN OF RAD53 COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Byeon, I.-J.L, Liao, H, Yongkiettrakul, S, Tsai, M.-D. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides.
J.Mol.Biol., 302, 2000
|
|
1FHQ
 
 | |
4PY4
 
 | |
7E6Q
 
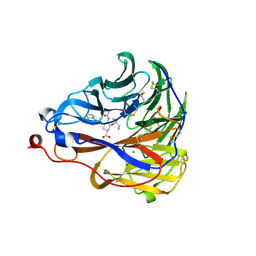 | | Crystal structure of influenza A virus neuraminidase N5 complexed with 4'-phenyl-1,2,3-triazolylated oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-acetamido-3-pentan-3-yloxy-5-(4-phenyl-1,2,3-triazol-1-yl)cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, P.F, Babayemi, O.O, Li, C.N, Fu, L.F, Zhang, S.S, Qi, J.X, Lv, X, Li, X.B. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of 5'-substituted 1,2,3-triazolylated oseltamivir derivatives as potent influenza neuraminidase inhibitors.
Rsc Adv, 11, 2021
|
|
7EDO
 
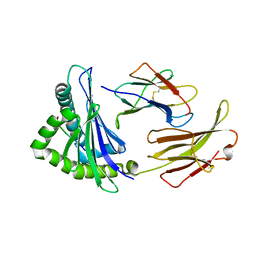 | | First insight into marsupial MHC I peptide presentation: immune features of lower mammals paralleled with bats | | Descriptor: | Beta-2-microglobulin, CYS-ASN-VAL-THR-LEU-ASN-TYR-PRO, MHC class I antigen | | Authors: | Wang, P.Y, Yue, C, Lu, D, Liu, K.F, Liu, S, Yao, S.J, Chai, Y, Qi, J.X, Lou, Y.L, Sun, Z.Y, Gao, G.F, Liu, W.J. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptide Presentations of Marsupial MHC Class I Visualize Immune Features of Lower Mammals Paralleled with Bats.
J Immunol., 207, 2021
|
|
7RX6
 
 | |
7RX7
 
 | |
7RX8
 
 | |
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
9L4B
 
 | | Kinase domain of ATR bound with RP-3500 | | Descriptor: | RP-3500, Serine/threonine-protein kinase ATR | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L46
 
 | | ATR-ATRIP-bound with AMP-PNP | | Descriptor: | ATR-interacting protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ATR | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.29 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L4C
 
 | | ATR Spiral -ATRIP bound with RP-3500 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, ZINC ION | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L40
 
 | | kinase of ATR bound VE-822 state | | Descriptor: | Serine/threonine-protein kinase ATR, VE-822 | | Authors: | Wang, G. | | Deposit date: | 2024-12-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L43
 
 | | ATR Spiral -ATRIP bound with VE-822 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, ZINC ION | | Authors: | Wang, G. | | Deposit date: | 2024-12-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L4F
 
 | | ATR-ATRIP bound with ATPgammaS | | Descriptor: | ATR-interacting protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ATR | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.22 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L4D
 
 | | ATR-ATRIP bound with RP-3500 | | Descriptor: | ATR-interacting protein, RP-3500, Serine/threonine-protein kinase ATR, ... | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L45
 
 | | ATR-ATRIP bound with VE-822 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, VE-822, ... | | Authors: | Wang, G. | | Deposit date: | 2024-12-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
8ROY
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
