5F8E
 
 | | Rv2258c-SAH | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5HS9
 
 | |
5HS7
 
 | |
5F8F
 
 | | Rv2258c-SFG | | Descriptor: | GLYCEROL, Methyltransferase, SINEFUNGIN | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5F8C
 
 | | Rv2258c-unbound | | Descriptor: | GLYCEROL, Methyltransferase | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5HS8
 
 | |
5GHV
 
 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-({1-[2-({3,5-dimethyl-4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]-4-methyl-1H-pyrrol-3-yl}methyl)azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
7UX9
 
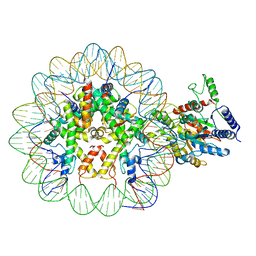 | | Arabidopsis DDM1 bound to nucleosome (H2A.W, H2B, H3.3, H4, with 147 bp DNA) | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Ipsaro, J.J, Adams, D.W, Joshua-Tor, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin remodeling of histone H3 variants by DDM1 underlies epigenetic inheritance of DNA methylation.
Cell, 186, 2023
|
|
7JTP
 
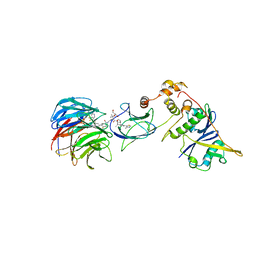 | | Crystal structure of Protac MS67 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | Elongin-B, Elongin-C, GLYCEROL, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
7JTO
 
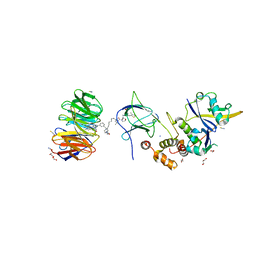 | | Crystal structure of Protac MS33 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(11-{[2-(4-{[4'-(4-methylpiperazin-1-yl)-3'-{[6-oxo-4-(trifluoromethyl)-5,6-dihydropyridine-3-carbonyl]amino}[1,1'-biphenyl]-3-yl]methyl}piperazin-1-yl)ethyl]amino}-11-oxoundecanoyl)-L-valyl-(4R)-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, Elongin-B, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
5ENP
 
 | | MBX2931 bound structure of bacterial efflux pump. | | Descriptor: | 6-[2-(3,4-dimethoxyphenyl)ethylsulfanyl]-8-[4-(2-methoxyethyl)piperazin-1-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENQ
 
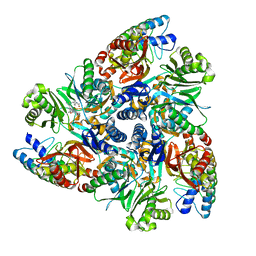 | | MBX3132 bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, ~{N}-[4-[2-[[5-cyano-8-[(2~{S},6~{R})-2,6-dimethylmorpholin-4-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridin-6-yl]sulfanyl]ethyl]phenyl]ethanamide | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENT
 
 | | Minocycline bound structure of bacterial efflux pump. | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENR
 
 | | MBX3135 bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, ~{N}-[4-[2-[[5-cyano-8-[(2~{S},6~{S})-2,6-dimethylmorpholin-4-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridin-6-yl]sulfanyl]ethyl]phenyl]prop-2-enamide | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENO
 
 | | MBX2319 bound structure of bacterial efflux pump. | | Descriptor: | 3,3-dimethyl-8-morpholin-4-yl-6-(2-phenylethylsulfanyl)-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENS
 
 | | Rhodamine bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, RHODAMINE 6G | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EN5
 
 | | Apo structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4O9D
 
 | | Structure of Dos1 propeller | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Rik1-associated factor 1 | | Authors: | Kuscu, C, Schalch, T, Joshua-Tor, L. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRL4-like Clr4 complex in Schizosaccharomyces pombe depends on an exposed surface of Dos1 for heterochromatin silencing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5GTO
 
 | | Human PPARgamma ligand binding dmain complexed with S35 | | Descriptor: | 2-[4-[5-[(1~{S})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Jang, J.Y, Suh, S.W. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
5GTN
 
 | | Human PPARgamma ligand binding dmain complexed with R35 | | Descriptor: | 2-[4-[5-[(1~{R})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Suh, S.W. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
4PKL
 
 | | Bromodomain of Trypanosoma brucei BDF2 With IBET-151 | | Descriptor: | 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, Bromodomain factor 2, CHLORIDE ION, ... | | Authors: | Debler, E.W. | | Deposit date: | 2014-05-15 | | Release date: | 2015-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Bromodomain Proteins Contribute to Maintenance of Bloodstream Form Stage Identity in the African Trypanosome.
Plos Biol., 13, 2015
|
|
5GTP
 
 | |
5XIX
 
 | | The canonical domain of human asparaginyl-tRNA synthetase | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, GLYCEROL | | Authors: | Park, J.S, Han, B.W. | | Deposit date: | 2017-04-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Unique N-terminal extension domain of human asparaginyl-tRNA synthetase elicits CCR3-mediated chemokine activity.
Int. J. Biol. Macromol., 120, 2018
|
|
6AFM
 
 | |
6AFO
 
 | |
