8R5N
 
 | | DTX1 WWE domain in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, E3 ubiquitin-protein ligase DTX1 | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8R6B
 
 | |
8R6A
 
 | |
8R7O
 
 | | HUWE1 WWE domain in complex with 2'F-ATP | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1, SODIUM ION, ... | | Authors: | Muenzker, L, Boettcher, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8RD1
 
 | | HUWE1 WWE domain in complex with compound 4 | | Descriptor: | 2-(2-hydroxy-2-oxoethyl)-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8RD0
 
 | | HUWE1 WWE domain in complex with compound 3 | | Descriptor: | (1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetic acid, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Muenzker, L, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
1CE2
 
 | | STRUCTURE OF DIFERRIC BUFFALO LACTOFERRIN AT 2.5A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (LACTOFERRIN) | | Authors: | Karthikeyan, S, Paramasivam, M, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of buffalo lactoferrin at 2.5 A resolution using crystals grown at 303 K shows different orientations of the N and C lobes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1DPJ
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH IA3 PEPTIDE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEINASE A, PROTEINASE INHIBITOR IA3 PEPTIDE, ... | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-27 | | Release date: | 2000-05-03 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1DP5
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT INHIBITOR | | Descriptor: | PROTEINASE A, PROTEINASE INHIBITOR IA3, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-23 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
5JN8
 
 | | Crystal Structure for the complex of human carbonic anhydrase IV and acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5JN9
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and ethoxyzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5IPZ
 
 | | Crystal structure of human carbonic anhydrase isozyme IV with 5-(2-amino-1,3-thiazol-4-yl)-2-chlorobenzenesulfonamide | | Descriptor: | 5-(2-amino-1,3-thiazol-4-yl)-2-chlorobenzene-1-sulfonamide, Carbonic anhydrase 4, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-03-10 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5JNC
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and 4-aminomethylbenzene sulfonamide | | Descriptor: | 4-(aminomethyl)benzene-1-sulfonamide, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5JNA
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and topiramate | | Descriptor: | ACETATE ION, Carbonic anhydrase 4, GLYCEROL, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
6KFE
 
 | |
6KFB
 
 | | Hydroxynitrile lyase from the millipede, Chamberlinius hualienensis bound with thiocyanate | | Descriptor: | Hydroxynitrile lyase, THIOCYANATE ION, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Motojima, F, Izumi, A, Asano, Y. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | R-hydroxynitrile lyase from the cyanogenic millipede, Chamberlinius hualienensis-A new entry to the carrier protein family Lipocalines.
Febs J., 288, 2021
|
|
1BKA
 
 | | OXALATE-SUBSTITUTED DIFERRIC LACTOFERRIN | | Descriptor: | FE (III) ION, LACTOFERRIN, OXALATE ION | | Authors: | Baker, H.M, Smith, C.A, Baker, E.N. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anion binding by transferrins: importance of second-shell effects revealed by the crystal structure of oxalate-substituted diferric lactoferrin.
Biochemistry, 35, 1996
|
|
5NCR
 
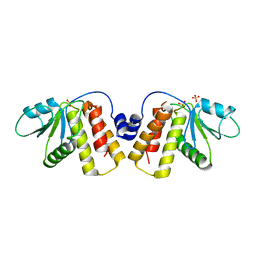 | | OH1 from the Orf virus: a tyrosine phosphatase that displays distinct structural features and triple substrate specificity | | Descriptor: | PHOSPHATE ION, SULFATE ION, tyrosine phosphatase | | Authors: | Segovia, D, Haouz, A, Berois, M, Villarino, A, Andre-Leroux, G. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | OH1 from Orf Virus: A New Tyrosine Phosphatase that Displays Distinct Structural Features and Triple Substrate Specificity.
J. Mol. Biol., 429, 2017
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
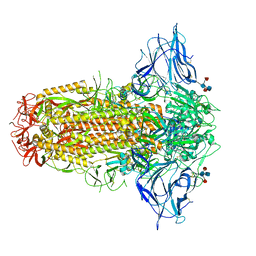 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7M78
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M75
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M76
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7Z3E
 
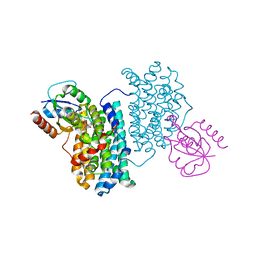 | | XFEL structure of Class Ib ribonucleotide reductase dimanganese(II) NrdF in complex with hydroquinone NrdI from Bacillus cereus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, MANGANESE (II) ION, Protein NrdI, ... | | Authors: | John, J, Lebrette, H, Aurelius, O, Hogbom, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-controlled reorganization and flavin strain within the ribonucleotide reductase R2b-NrdI complex monitored by serial femtosecond crystallography.
Elife, 11, 2022
|
|
7Z3D
 
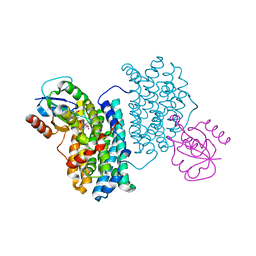 | | XFEL structure of Class Ib ribonucleotide reductase dimanganese(II) NrdF in complex with oxidized NrdI from Bacillus cereus | | Descriptor: | FLAVIN MONONUCLEOTIDE, MANGANESE (II) ION, Protein NrdI, ... | | Authors: | John, J, Lebrette, H, Aurelius, O, Hogbom, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-controlled reorganization and flavin strain within the ribonucleotide reductase R2b-NrdI complex monitored by serial femtosecond crystallography.
Elife, 11, 2022
|
|
