5TUX
 
 | | crystal structure and light induced structural changes in orange carotenoid protein bound with echinenone | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YJW
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with the competitive inhibitor, stigmatellin. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5TUW
 
 | | Crystal structure of Orange Carotenoid Protein with partial loss of 3'OH Echinenone chromophore | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, GLYCEROL, Orange carotenoid-binding protein | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMB
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7CE4
 
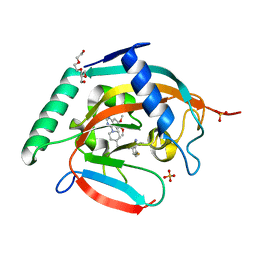 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
9BA4
 
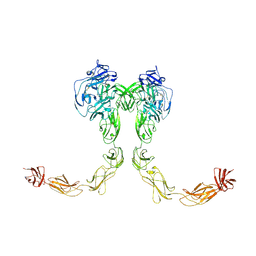 | | Full-length cross-linked Contactin 2 (CNTN2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2 | | Authors: | Liu, J.L, Fan, S.F, Ren, G.R, Rudenko, G.R. | | Deposit date: | 2024-04-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of contactin 2 homophilic interaction.
Structure, 2024
|
|
9BA5
 
 | |
6JNO
 
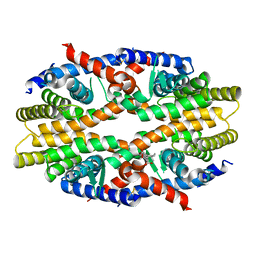 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
8JZD
 
 | | Crystal structure of Escherichia coli NarJ in complex with the signal peptide of E. coli NarG | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone NarJ, Respiratory nitrate reductase 1 alpha chain | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
4XVE
 
 | |
4BGD
 
 | | Crystal structure of Brr2 in complex with the Jab1/MPN domain of Prp8 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, T.H.D, Li, J, Nagai, K. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Brr2-Prp8 Interactions and Implications for U5 Snrnp Biogenesis and the Spliceosome Active Site
Structure, 21, 2013
|
|
5F9E
 
 | | Structure of Protein Kinase C theta with compound 10: 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one | | Descriptor: | 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one, Protein kinase C theta type | | Authors: | Klein, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of 1,7-disubstituted-2,2-dimethyl-2,3-dihydroquinazolin-4(1H)-ones as potent and selective PKC theta inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
2RQL
 
 | |
8JZC
 
 | | Crystal structure of Geobacillus stearothermophilus NarJ | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
2G5R
 
 | | Crystal structure of Siglec-7 in complex with methyl-9-(aminooxalyl-amino)-9-deoxyNeu5Ac (oxamido-Neu5Ac) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Sialic acid-binding Ig-like lectin 7, ... | | Authors: | Attrill, H, Crocker, P.R, van Aalten, D.M. | | Deposit date: | 2006-02-23 | | Release date: | 2006-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of siglec-7 in complex with sialosides: leads for rational structure-based inhibitor design.
Biochem.J., 397, 2006
|
|
3RXB
 
 | | Crystal structure of Trypsin complexed with 4-guanidinobutanoic acid | | Descriptor: | 4-carbamimidamidobutanoic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXP
 
 | | Crystal structure of Trypsin complexed with (1,5-dimethylpyrazol-3-yl)methanamine | | Descriptor: | 1-(1,5-dimethyl-1H-pyrazol-3-yl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXD
 
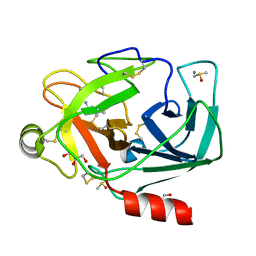 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamine | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXM
 
 | | Crystal structure of Trypsin complexed with [2-(2-thienyl)thiazol-4-yl]methanamine | | Descriptor: | 1-[2-(thiophen-2-yl)-1,3-thiazol-4-yl]methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXH
 
 | | Crystal structure of Trypsin complexed with 2-(1H-imidazol-4-yl)ethanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXF
 
 | | Crystal structure of Trypsin complexed with 4-aminopyridine | | Descriptor: | 4-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
5ZFP
 
 | |
3RXG
 
 | | Crystal structure of Trypsin complexed with 4-aminocyclohexanol | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXT
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamin (F04 and F03, cocktail experiment) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
