6VAH
 
 | | Crystal structure of human TEAD2 transcription factor in complex with Flufenamic acid derivative | | Descriptor: | 2-fluoro-6-[(3-hexylphenyl)amino]benzoic acid, Transcriptional enhancer factor TEF-4, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Dong, A, Li, Y, Melin, L, Gagnon, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of human TEAD2 transcription factor in complex with Flufenamic acid derivative
to be published
|
|
5BUP
 
 | | Crystal structure of the ZP-C domain of mouse ZP2 | | Descriptor: | ACETATE ION, Zona pellucida sperm-binding protein 2 | | Authors: | Nishimura, K, Jovine, L. | | Deposit date: | 2015-06-04 | | Release date: | 2016-01-27 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | A structured interdomain linker directs self-polymerization of human uromodulin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2X24
 
 | | bovine ACC2 CT domain in complex with inhibitor | | Descriptor: | ACETYL-COA CARBOXYLASE, TERT-BUTYL [(TRANS-4-{[({2-[4-(AMINOMETHYL)PHENYL]QUINOLIN-4-YL}CARBONYL)AMINO]METHYL}CYCLOHEXYL)METHYL]CARBAMATE | | Authors: | Oster, L, Folmer, R, Blaho, S, Wiberg, F, Hallberg, K. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of Small Molecule Inhibitors of Acetyl-Coa Carboxylase 1 and 2 Showing Reduction of Hepatic Malonyl-Coa Levels in Vivo in Obese Zucker Rats.
Bioorg.Med.Chem., 19, 2011
|
|
8GRQ
 
 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | Descriptor: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
2VM6
 
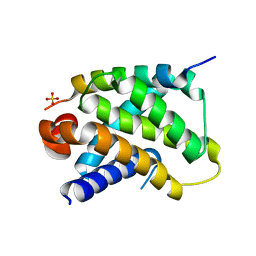 | | Human Bcl2-A1 in complex with Bim-BH3 peptide | | Descriptor: | BCL-2-LIKE PROTEIN 11, BCL-2-RELATED PROTEIN A1, SULFATE ION | | Authors: | Herman, M.D, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-23 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Completing the Family Portrait of the Anti- Apoptotic Bcl-2 Proteins: Crystal Structure of Human Bfl-1 in Complex with Bim.
FEBS Lett., 582, 2008
|
|
2WPO
 
 | | HCMV protease inhibitor complex | | Descriptor: | (2S)-2-(3,3-dimethylbutanoylamino)-N-[(2S)-1-[[(2S,3S)-3-hydroxy-4-[(4-iodophenyl)methylamino]-4-oxo-butan-2-yl]amino]- 1,4-dioxo-4-pyrrol-1-yl-butan-2-yl]-3,3-dimethyl-butanamide, HUMAN CYTOMEGALOVIRUS PROTEASE | | Authors: | Tong, L, Qian, C, Massariol, M.-J, Deziel, R, Yoakim, C, Lagace, L. | | Deposit date: | 1998-08-04 | | Release date: | 1999-08-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conserved mode of peptidomimetic inhibition and substrate recognition of human cytomegalovirus protease.
Nat.Struct.Biol., 5, 1998
|
|
1PDC
 
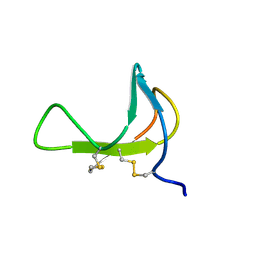 | |
5GI4
 
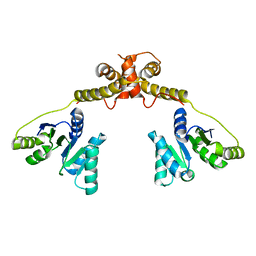 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
7TTM
 
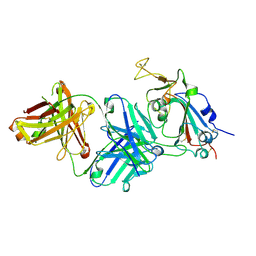 | |
7TTX
 
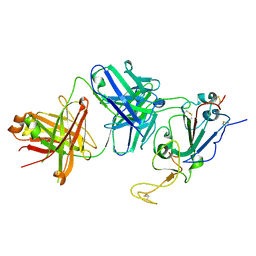 | |
6VP0
 
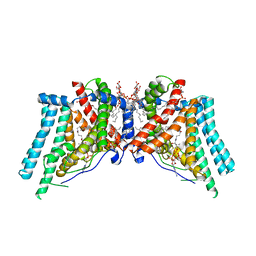 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
3B9O
 
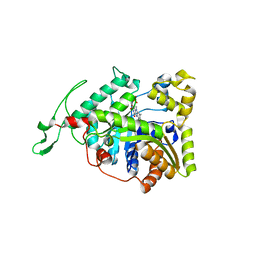 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
6W75
 
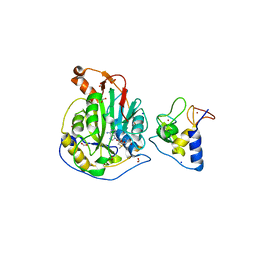 | | 1.95 Angstrom Resolution Crystal Structure of NSP10 - NSP16 Complex from SARS-CoV-2 | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6VXG
 
 | | Structure of the C-terminal Domain of RAGE and Its Inhibitor | | Descriptor: | Advanced glycosylation end product-specific receptor, N-(4-{7-cyano-4-[(morpholin-4-yl)methyl]quinolin-2-yl}phenyl)acetamide | | Authors: | Ramirez, L, Shekhtman, A. | | Deposit date: | 2020-02-21 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small-molecule antagonism of the interaction of the RAGE cytoplasmic domain with DIAPH1 reduces diabetic complications in mice.
Sci Transl Med, 13, 2021
|
|
2WKS
 
 | | Structure of Helicobacter pylori Type II Dehydroquinase with a new carbasugar-thiophene inhibitor. | | Descriptor: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Guardado-Calvo, P, Llamas-Saiz, A.L, Prazeres, V.F.V, Tizon, L, Castedo, L, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues.
J. Med. Chem., 53, 2010
|
|
1PVL
 
 | | STRUCTURE OF THE PANTON-VALENTINE LEUCOCIDIN F COMPONENT FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LEUCOCIDIN | | Authors: | Pedelacq, J.D, Mourey, L, Maveyraud, L, Prevost, G, Samama, J.P. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a Staphylococcus aureus leucocidin component (LukF-PV) reveals the fold of the water-soluble species of a family of transmembrane pore-forming toxins.
Structure Fold.Des., 7, 1999
|
|
6Z1C
 
 | | Crystal structure of Arabidopsis thaliana CK2-alpha-1 in complex with TTP-22 | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha-1 | | Authors: | Demulder, M, Loris, R, De Veylder, L. | | Deposit date: | 2020-05-13 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis casein kinase 2 triggers stem cell exhaustion under Al toxicity and phosphate deficiency through activating the DNA damage response pathway.
Plant Cell, 33, 2021
|
|
2V9Y
 
 | | Human aminoimidazole ribonucleotide synthetase | | Descriptor: | PHOSPHORIBOSYLFORMYLGLYCINAMIDINE CYCLO-LIGASE, SULFATE ION | | Authors: | Welin, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Herman, M.D, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies of Tri-Functional Human Gart.
Nucleic Acids Res., 38, 2010
|
|
8GRM
 
 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | Descriptor: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
7TTY
 
 | |
3B9N
 
 | | Crystal structure of long-chain alkane monooxygenase (LadA) | | Descriptor: | Alkane monooxygenase | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
7V3Z
 
 | | Structure of cannabinoid receptor type 1(CB1) | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, CHOLESTEROL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liu, Z.J, Shen, L, Hua, T, Yao, D.Q, Wu, L.J. | | Deposit date: | 2021-08-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | A Genetically Encoded F-19 NMR Probe Reveals the Allosteric Modulation Mechanism of Cannabinoid Receptor 1.
J.Am.Chem.Soc., 143, 2021
|
|
1GNO
 
 | | HIV-1 PROTEASE (WILD TYPE) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
1NTO
 
 | | N249Y MUTANT OF ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS-MONOCLINIC CRYSTAL FORM | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-01-30 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural study of a single-point mutant of Sulfolobus solfataricus alcohol dehydrogenase with enhanced activity.
Febs Lett., 539, 2003
|
|
4AQL
 
 | | HUMAN GUANINE DEAMINASE IN COMPLEX WITH VALACYCLOVIR | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl L-valinate, GUANINE DEAMINASE, ZINC ION | | Authors: | Welin, M, Egeblad, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, Weigelt, J, Nordlund, P. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Pan-Pathway Based Interaction Profiling of Fda-Approved Nucleoside and Nucleobase Analogs with Enzymes of the Human Nucleotide Metabolism.
Plos One, 7, 2012
|
|
