8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8DGF
 
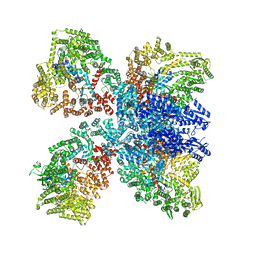 | | Avs4 bound to phage PhiV-1 portal | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding protein Avs4, MAGNESIUM ION, ... | | Authors: | Wilkinson, M.E, Gao, L, Strecker, J, Makarova, K.S, Macrae, R.K, Koonin, E.V, Zhang, F. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Prokaryotic innate immunity through pattern recognition of conserved viral proteins.
Science, 377, 2022
|
|
8DGC
 
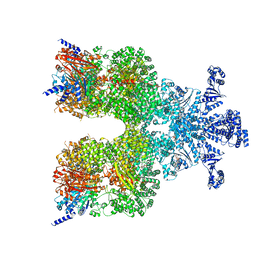 | | Avs3 bound to phage PhiV-1 terminase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SeAvs3, ... | | Authors: | Wilkinson, M.E, Gao, L, Strecker, J, Makarova, K.S, Macrae, R.K, Koonin, E.V, Zhang, F. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Prokaryotic innate immunity through pattern recognition of conserved viral proteins.
Science, 377, 2022
|
|
8DMB
 
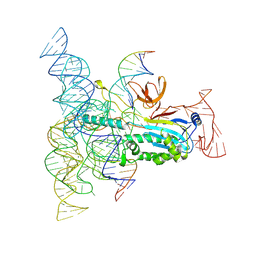 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | Descriptor: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | Authors: | Seiichi, H, Kappel, K, Zhang, F. | | Deposit date: | 2022-07-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
8EEX
 
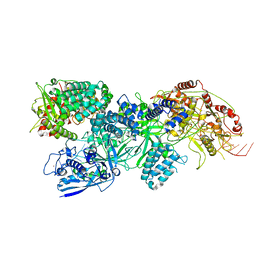 | | Cas7-11 in complex with Csx29 | | Descriptor: | Cas7-11, Csx29, ZINC ION, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
8EEY
 
 | | Cas7-11 in complex with DR-mismatched target RNA, Csx29 and Csx30 | | Descriptor: | Cas7-11, Csx29, Csx30, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
4U63
 
 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
8E4L
 
 | | The open state mouse TRPM8 structure in complex with the cooling agonist C3, AITC, and PI(4,5)P2 | | Descriptor: | 3-isothiocyanatoprop-1-ene, CALCIUM ION, Transient receptor potential cation channel subfamily M member 8, ... | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4O
 
 | | The closed C1-state mouse TRPM8 structure in complex with putative PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4N
 
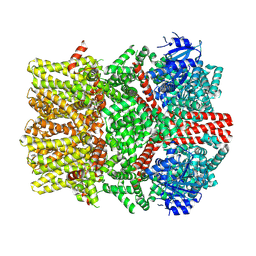 | | The closed C1-state mouse TRPM8 structure in complex with PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4Q
 
 | | The closed C0-state flycatcher TRPM8 structure in complex with PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4P
 
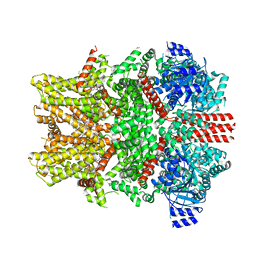 | | Mouse TRPM8 structure determined in the ligand- and PI(4,5)P2-free condition, Class I , C0 state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4M
 
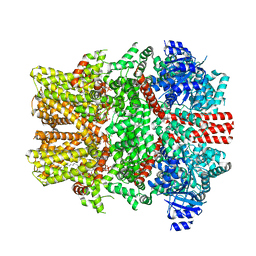 | | The intermediate C2-state mouse TRPM8 structure in complex with the cooling agonist C3 and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
6LQX
 
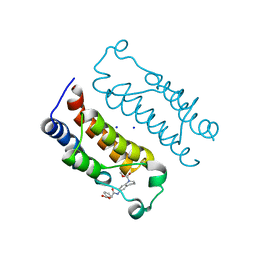 | | Crystal structure of the CBP bromodomain in complex with small molecule LC-CPin7 | | Descriptor: | (1~{S},6~{R})-6-[(1-methoxycarbonyl-3,4-dihydro-2~{H}-quinolin-6-yl)carbamoyl]cyclohex-3-ene-1-carboxylic acid, CREB-binding protein, SODIUM ION | | Authors: | Chen, Y, Zhang, F, Sun, Z, Bi, X, Luo, C. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Design, synthesis and biological evaluation of novel small molecule inhibitor of the CBP bromodomain with possible anti-leukemia effects
To Be Published
|
|
4END
 
 | | Crystal structure of anti-HIV actinohivin in complex with alpha-1,2-mannobiose (P 2 21 21 form) | | Descriptor: | ACETONITRILE, Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Matured structure of anti-HIV lectin actinohivin in complex with 1,2-mannobiose
To be Published
|
|
8T2I
 
 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | Descriptor: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | Authors: | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
4G1R
 
 | | Crystal structure of anti-HIV actinohivin in complex with alphs-1,2-mannobiose (Form II) | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-07-11 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Matured structure of anti-HIV lectin actinohivin in complex with alpha-1,2-mannobiose
To be Published
|
|
8FC7
 
 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK2798745 | | Descriptor: | 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FC9
 
 | | Cryo-EM structure of the human TRPV4 - RhoA, apo condition | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FC8
 
 | | Cryo-EM structure of the human TRPV4 in complex with GSK1016790A | | Descriptor: | CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FCA
 
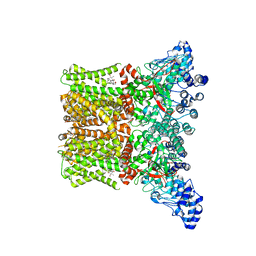 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with 4alpha-Phorbol 12,13-didecanoate | | Descriptor: | (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
1D8K
 
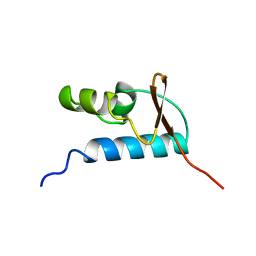 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1D8J
 
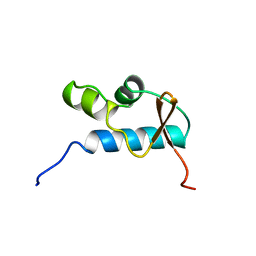 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
5UF6
 
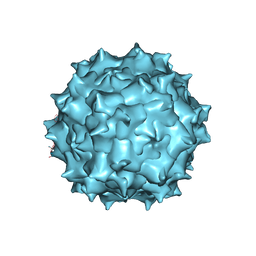 | | The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, capsid protein VP1 | | Authors: | Xie, Q, Spear, J.M, Noble, A.J, Sousa, D.R, Meyer, N.L, Davulcu, O, Zhang, F, Linhardt, R.J, Stagg, S.M, Chapman, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The 2.8 angstrom Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparinoid Pentasaccharide.
Mol Ther Methods Clin Dev, 5, 2017
|
|
