5KDM
 
 | |
4FWE
 
 | | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWF
 
 | | Complex structure of LSD2/AOF1/KDM1b with H3K4 mimic | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.1, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWJ
 
 | | Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, PHOSPHATE ION, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
6JDS
 
 | |
6JDU
 
 | | Crystal structure of PRRSV nsp10 (helicase) | | Descriptor: | CALCIUM ION, PP1b, ZINC ION | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2019-02-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
5EHP
 
 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP836 | | Descriptor: | 5-[2,3-bis(chloranyl)phenyl]-2-[(3~{R},5~{S})-3,5-dimethylpiperazin-1-yl]pyrimidin-4-amine, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor.
J.Med.Chem., 59, 2016
|
|
5EHR
 
 | |
6K9N
 
 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6KBE
 
 | | Structure of Deubiquitinase | | Descriptor: | Polyubiquitin-C, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6K9P
 
 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
8JWN
 
 | | Crystal structure of AKRtyl-NADPH complex | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWO
 
 | |
8JWL
 
 | |
8JWK
 
 | | The second purified state crystal structure of AKRtyl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWM
 
 | | Crystal structure of AKRtyl-NADP-tylosin complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TYLOSIN | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
6JUF
 
 | | SspB crystal structure | | Descriptor: | SspB protein | | Authors: | Liqiong, L, Yubing, Z. | | Deposit date: | 2019-04-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
8K4Q
 
 | |
6PLK
 
 | |
4NRP
 
 | |
4NRM
 
 | |
4NRQ
 
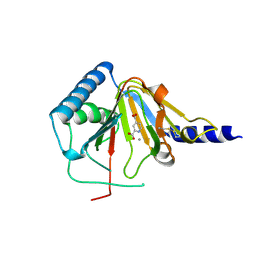 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | Descriptor: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | Authors: | Feng, C, Chen, Z, Liu, Y. | | Deposit date: | 2013-11-27 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
4NRO
 
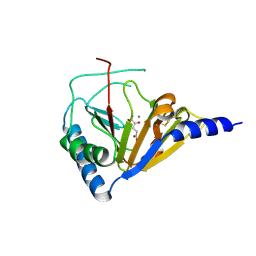 | |
6JIV
 
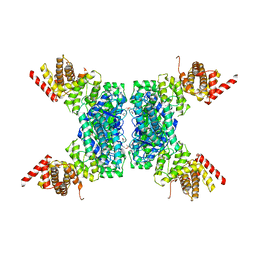 | | SspE crystal structure | | Descriptor: | SspE protein | | Authors: | Bing, Y.Z, Yang, H.G. | | Deposit date: | 2019-02-23 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
4O7X
 
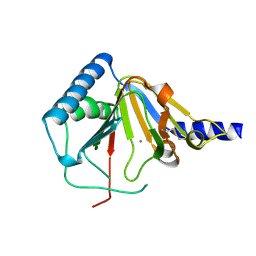 | |
