5JWS
 
 | | T4 Lysozyme L99A with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWW
 
 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
1TKU
 
 | | Crystal Structure of 3,4-Dihydroxy-2-butanone 4-phosphate Synthase of Candida albicans in complex with Ribulose-5-phosphate | | Descriptor: | 3,4-Dihydroxy-2-butanone 4-phosphate Synthase, RIBULOSE-5-PHOSPHATE | | Authors: | Echt, S, Bauer, S, Steinbacher, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-06-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans.
J.Mol.Biol., 341, 2004
|
|
4JM6
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2,4-diaminopyrimidine | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
1TKS
 
 | | Crystal structure of 3,4-Dihydroxy-2-butanone 4-phosphate Synthase of Candida albicans | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase | | Authors: | Echt, S, Bauer, S, Steinbacher, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-06-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans.
J.Mol.Biol., 341, 2004
|
|
5MRN
 
 | | Arabidopsis thaliana IspD Glu258Ala Mutant | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRQ
 
 | | Arabidopsis thaliana IspD Asp262Ala Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRM
 
 | | Arabidopsis thaliana IspD in complex with Isoxazole (4) | | Descriptor: | 2,4-bis(bromanyl)-6-[3-(trifluoromethyl)-1,2-oxazol-5-yl]phenol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRO
 
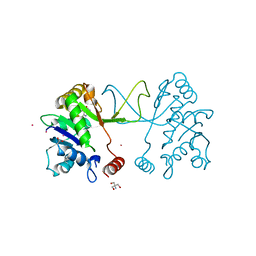 | | Arabidopsis thaliana IspD Glu258Ala mutant in complex with Azolopyrimidine (1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRP
 
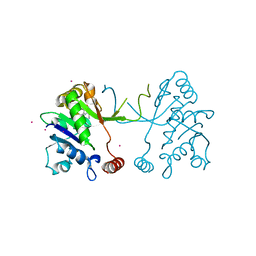 | | Arabidopsis thaliana IspD Glu258Ala mutant in complex with Azolopyrimidine (2) | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, 5-chloro-7-hydroxy-6-(phenylmethyl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
2F59
 
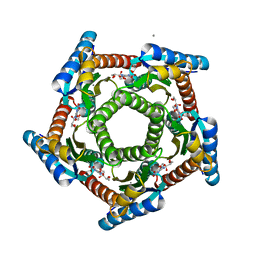 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
4NAL
 
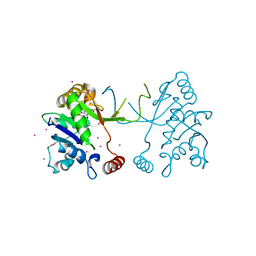 | | Arabidopsis thaliana IspD in complex with tribromodichloro-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAI
 
 | | Arabidopsis thaliana IspD apo | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAK
 
 | | Arabidopsis thaliana IspD in complex with pentabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAN
 
 | | Arabidopsis thaliana IspD in complex with tetrabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7M2E
 
 | | Crystal structure of BPTF bromodomain in complex with CB02-092 | | Descriptor: | 4-chloro-5-{4-[2-(dimethylamino)ethyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Nithianantham, S, Fischer, M. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition
J.Med.Chem., 64, 2021
|
|
7RN2
 
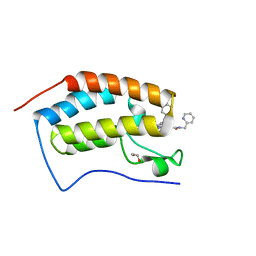 | | Crystal structure of the first bromodomain of human BRD4 in complex with SJ001010551-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(6S,10S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-[(pyridin-2-yl)methyl]acetamide, Bromodomain-containing protein 4 | | Authors: | Stachowski, T.R, Fischer, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | From PROTAC to inhibitor: Structure-guided discovery of potent and orally bioavailable BET inhibitors.
Eur.J.Med.Chem., 251, 2023
|
|
7RMD
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SJ001011461-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(6S,10R)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(1,3,4-thiadiazol-2-yl)acetamide, Bromodomain-containing protein 4 | | Authors: | Stachowski, T.R, Fischer, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | From PROTAC to inhibitor: Structure-guided discovery of potent and orally bioavailable BET inhibitors.
Eur.J.Med.Chem., 251, 2023
|
|
7S8Z
 
 | |
7S9G
 
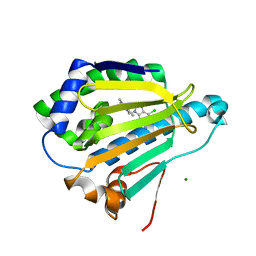 | | Room-temperature Human Hsp90a-NTD bound to BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Stachowski, T.R, Vanarotti, M, Fischer, M. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S9F
 
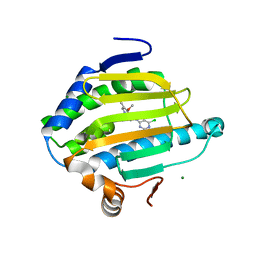 | | Cryogenic Human Hsp90a-NTD bound to BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Stachowski, T.R, Vanarotti, M, Seetharaman, J, Fischer, M. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S95
 
 | | Room-temperature Human Hsp90a-NTD bound to adenine | | Descriptor: | ADENINE, Heat shock protein HSP 90-alpha | | Authors: | Stachowski, T.R, Vanarotti, M, Lopez, K, Fischer, M. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S9H
 
 | | Cryogenic Human Hsp90a-NTD bound to EC144 | | Descriptor: | 5-{2-amino-4-chloro-7-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-7H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-methylpent-4-yn-2 -ol, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Stachowski, T.R, Vanarotti, M, Seetharaman, J, Fischer, M. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S9I
 
 | | Room-temperature Human Hsp90a-NTD bound to EC144 | | Descriptor: | 5-{2-amino-4-chloro-7-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-7H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-methylpent-4-yn-2 -ol, Heat shock protein HSP 90-alpha | | Authors: | Stachowski, T.R, Vanarotti, M, Fischer, M. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S90
 
 | | Cryogenic Human Hsp90a-NTD bound to adenine | | Descriptor: | ADENINE, Heat shock protein HSP 90-alpha | | Authors: | Stachowski, T.R, Vanarotti, M, Lopez, K, Fischer, M. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
