5EDP
 
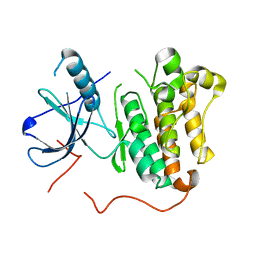 | | EGFR kinase (T790M/L858R) apo | | Descriptor: | Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 4-Aminoindazolyl-dihydrofuro[3,4-d]pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EDQ
 
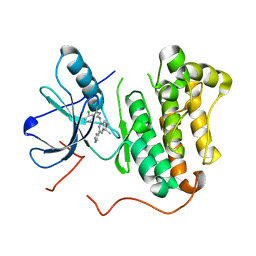 | | EGFR kinase (T790M/L858R) with inhibitor compound 15: ~{N}-(7-chloranyl-1~{H}-indazol-3-yl)-7,7-dimethyl-2-(1~{H}-pyrazol-4-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Descriptor: | Epidermal growth factor receptor, ~{N}-(7-chloranyl-1~{H}-indazol-3-yl)-7,7-dimethyl-2-(1~{H}-pyrazol-4-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 4-Aminoindazolyl-dihydrofuro[3,4-d]pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EM7
 
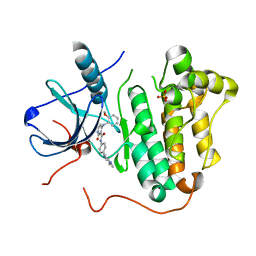 | |
5EM6
 
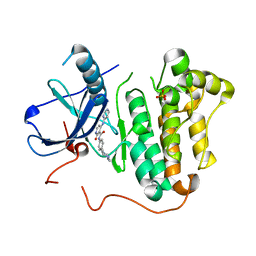 | |
5EM8
 
 | |
5EDR
 
 | | EGFR kinase (T790M/L858R) with inhibitor compound 27: ~{N}-(1~{H}-indazol-3-yl)-7,7-dimethyl-2-(2-methylpyrazol-3-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Descriptor: | Epidermal growth factor receptor, ~{N}-(1~{H}-indazol-3-yl)-7,7-dimethyl-2-(2-methylpyrazol-3-yl)-5~{H}-furo[3,4-d]pyrimidin-4-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 4-Aminoindazolyl-dihydrofuro[3,4-d]pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EM5
 
 | |
1M8R
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
5LUH
 
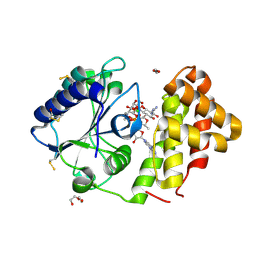 | | AadA E87Q in complex with ATP, calcium and streptomycin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Stern, A.L, Van der Verren, S.E, Selmer, M. | | Deposit date: | 2016-09-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J.Biol.Chem., 293, 2018
|
|
6ULU
 
 | | D104A/S128A S. typhimurium siroheme synthase | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-10-08 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
6KI0
 
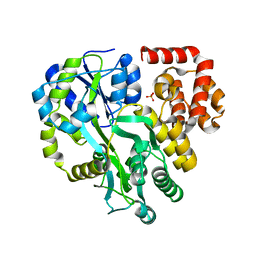 | | Crystal Structure of Human ASC-CARD | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, Z.H, Jin, T.C. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
6KUU
 
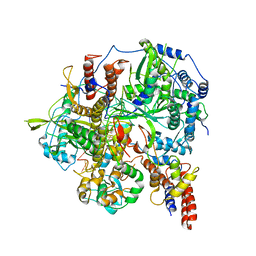 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
5LPA
 
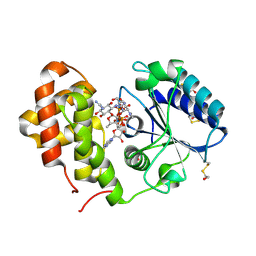 | | AadA E87Q in complex with ATP, calcium and dihydrostreptomycin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Stern, A.L, Van der Verren, S.E, Selmer, M. | | Deposit date: | 2016-08-12 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J.Biol.Chem., 293, 2018
|
|
6UBS
 
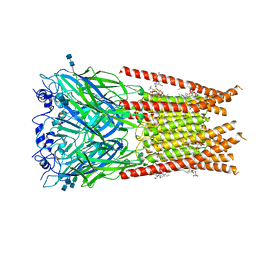 | | Full length Glycine receptor reconstituted in lipid nanodisc in Apo/Resting conformation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, ... | | Authors: | Kumar, A, Basak, S, Chakrapani, S. | | Deposit date: | 2019-09-12 | | Release date: | 2020-07-29 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
6VG0
 
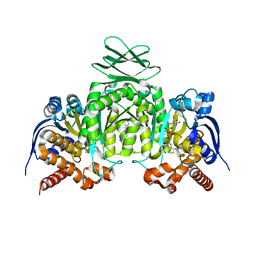 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC ISOCITRATE DEHYDROGENASE (IDH1) R132H MUTANT IN COMPLEX WITH NADPH and AGI-15056 | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~2~,N~4~-bis[(1R)-1-cyclopropylethyl]-6-[6-(trifluoromethyl)pyridin-2-yl]-1,3,5-triazine-2,4-diamine | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-01-07 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma.
Acs Med.Chem.Lett., 11, 2020
|
|
6L5D
 
 | |
5KRE
 
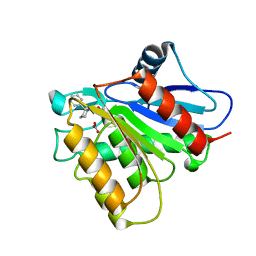 | | Covalent inhibitor of LYPLAL1 | | Descriptor: | (2~{R})-2-phenylpiperidine-1-carbaldehyde, Lysophospholipase-like protein 1, NITRATE ION | | Authors: | Pandit, J. | | Deposit date: | 2016-07-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Selective Covalent Inhibitor of Lysophospholipase-like 1 (LYPLAL1) as a Tool to Evaluate the Role of this Serine Hydrolase in Metabolism.
Acs Chem.Biol., 11, 2016
|
|
6DUN
 
 | | Crystal Structure Analysis of PIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, TRIHYDROXYARSENITE(III) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells.
Nat Commun, 9, 2018
|
|
3CAJ
 
 | | Crystal structure of the human carbonic anhydrase II in complex with ethoxzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, CHLORIDE ION, Carbonic anhydrase 2, ... | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2008-02-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Carbonic anhydrase inhibitors: the X-ray crystal structure of ethoxzolamide complexed to human isoform II reveals the importance of thr200 and gln92 for obtaining tight-binding inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7BPU
 
 | |
6KCV
 
 | | Structure of alginate lyase Aly36B mutant K143A/Y185A in complex with alginate tetrasaccharide | | Descriptor: | Alginate lyase, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Dong, F, Zhang, Y.Z, Chen, X.L. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
8TQM
 
 | |
8TS6
 
 | | Cyanophage A-1(L) portal | | Descriptor: | Portal protein | | Authors: | Yu, R.C, Li, Q, Zhou, C.Z. | | Deposit date: | 2023-08-11 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the intact tail machine of Anabaena myophage A-1(L).
Nat Commun, 15, 2024
|
|
6KZK
 
 | |
6WWZ
 
 | | Cryo-EM structure of the human chemokine receptor CCR6 in complex with CCL20 and a Go protein | | Descriptor: | C-C chemokine receptor type 6,C-C chemokine receptor type 6, C-C motif chemokine 20, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wasilko, D.J, Johnson, Z.L, Ammirati, M, Han, S, Wu, H. | | Deposit date: | 2020-05-09 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for chemokine receptor CCR6 activation by the endogenous protein ligand CCL20.
Nat Commun, 11, 2020
|
|
