5HRS
 
 | | HIV Integrase Catalytic Domain containing F185K + A124N + T125A mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5HRN
 
 | | HIV Integrase Catalytic Domain containing F185K mutation complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-23 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
6C0S
 
 | |
5CH8
 
 | | Crystal structure of MDLA N225Q mutant form Penicillium cyclopium | | Descriptor: | CHLORIDE ION, GLYCEROL, Mono- and diacylglycerol lipase, ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-07-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Lipase-Driven Epoxidation Is A Two-Stage Synergistic Process
ChemistrySelect, 4, 2016
|
|
8WRH
 
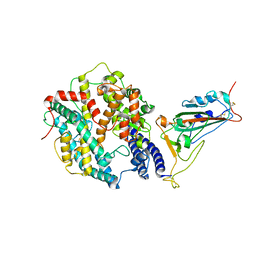 | | SARS-CoV-2 XBB.1.5.70 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRL
 
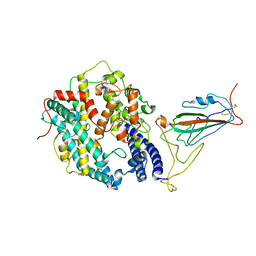 | | XBB.1.5 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRM
 
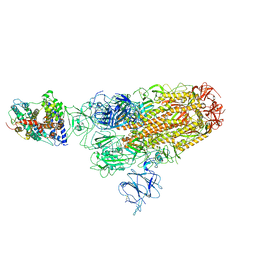 | | XBB.1.5 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRO
 
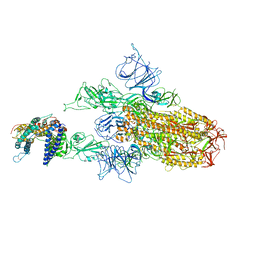 | | XBB.1.5.10 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein,Spike glycoprotein,Spike glycoprotein,Fusion protein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
9MZA
 
 | | Chemically Hijacked BCL6-TCIP3-p300 Complex | | Descriptor: | 1-{1-[5-({1-[5-chloro-4-({8-methoxy-1-methyl-3-[2-(methylamino)-2-oxoethoxy]-2-oxo-1,2-dihydroquinolin-6-yl}amino)pyrimidin-2-yl]piperidine-4-carbonyl}amino)pentanoyl]piperidin-4-yl}-3-[(6M)-7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, B-cell lymphoma 6 protein, Histone acetyltransferase p300 | | Authors: | Hinshaw, S.M, Gray, N.S, Nix, M.N, Gourisankar, S, Martinez, M, Crabtree, G.R. | | Deposit date: | 2025-01-22 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Bivalent Molecular Glue Linking Lysine Acetyltransferases to Oncogene-induced Cell Death.
Biorxiv, 2025
|
|
9M84
 
 | |
6KWX
 
 | | cryo-EM structure of human PA200 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, [(1~{S},2~{R},3~{R},4~{S},5~{S},6~{R})-2-[oxidanyl(phosphonooxy)phosphoryl]oxy-3,4,5,6-tetraphosphonooxy-cyclohexyl] phosphono hydrogen phosphate | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2019-09-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
6M3C
 
 | | hAPC-h1573 Fab complex | | Descriptor: | Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, h1573 Fab H chain, ... | | Authors: | Wang, X, Wang, D, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | Descriptor: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
1QT1
 
 | |
7PQX
 
 | |
7PSD
 
 | |
7PRO
 
 | |
7PRU
 
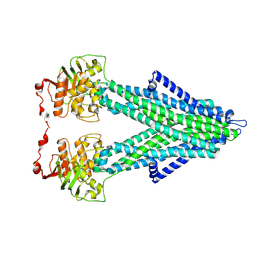 | |
7PR1
 
 | |
7KLZ
 
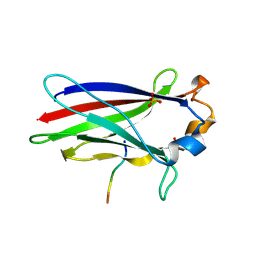 | |
5F5M
 
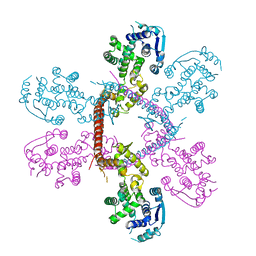 | | Crystal structure of Marburg virus nucleoprotein core domain | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
5F5O
 
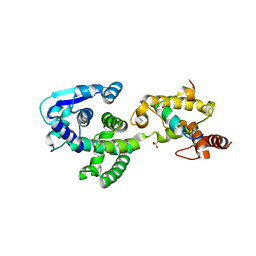 | | Crystal structure of Marburg virus nucleoprotein core domain bound to VP35 regulation peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35, SULFATE ION | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
6LNA
 
 | | YdiU complex with AMPNPP and Mn2+ | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
