7BZB
 
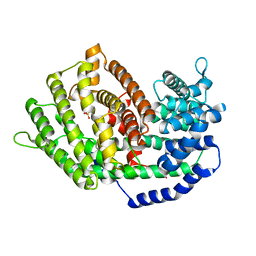 | |
7BZC
 
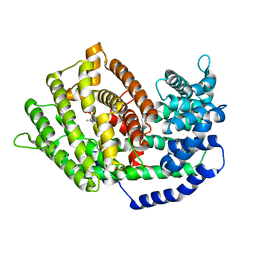 | | Crystal structure of plant sesterterpene synthase AtTPS18 complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, Terpenoid synthase 18 | | Authors: | Li, J.X, Wang, G.D, Zhang, P. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Molecular Basis for Sesterterpene Diversity Produced by Plant Terpene Synthases.
Plant Commun., 1, 2020
|
|
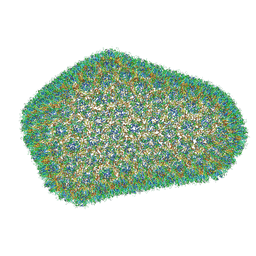
3J3Q
 
 | |
3J4F
 
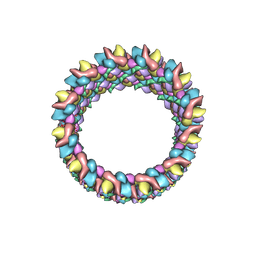 | | Structure of HIV-1 capsid protein by cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3JA6
 
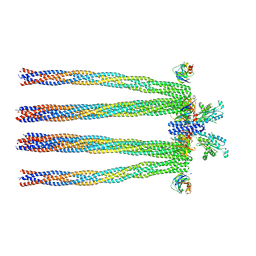 | | Cryo-electron Tomography and All-atom Molecular Dynamics Simulations Reveal a Novel Kinase Conformational Switch in Bacterial Chemotaxis Signaling | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein 2 | | Authors: | Cassidy, C.K, Himes, B.A, Alvarez, F.J, Ma, J, Zhao, G, Perilla, J.R, Schulten, K, Zhang, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Elife, 4, 2015
|
|
3H4P
 
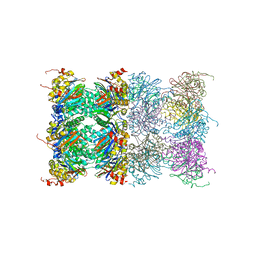 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H43
 
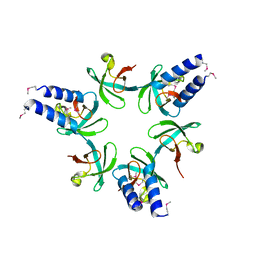 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | Descriptor: | Proteasome-activating nucleotidase | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
6XF7
 
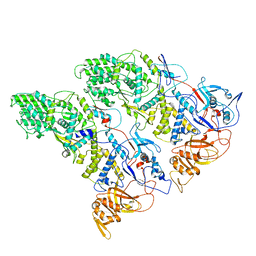 | | SLP | | Descriptor: | Lambda 1 protein | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
6Y9X
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,7) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9Z
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,9) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9V
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-8,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4Z7F
 
 | | Crystal structure of FolT bound with folic acid | | Descriptor: | FOLIC ACID, Folate ECF transporter | | Authors: | Zhao, Q, Wang, C.C, Wang, C.Y, Zhang, P. | | Deposit date: | 2015-04-07 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters
Nat Commun, 6, 2015
|
|
6Y9Y
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-7,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9W
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,8) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WJF
 
 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
4RFS
 
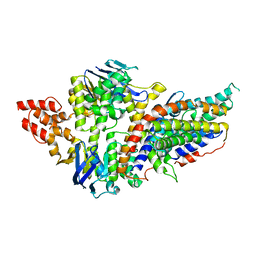 | | Structure of a pantothenate energy coupling factor transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Zhang, M, Bao, Z, Zhao, Q, Guo, H, Xu, K, Zhang, P. | | Deposit date: | 2014-09-27 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Structure of a pantothenate transporter and implications for ECF module sharing and energy coupling of group II ECF transporters.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7ZBT
 
 | | Subtomogram averaging of Rubisco from native Halothiobacillus carboxysomes | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small subunit | | Authors: | Ni, T, Zhu, Y, Yu, X, Sun, Y, Liu, L, Zhang, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-07-20 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and assembly of cargo Rubisco in two native alpha-carboxysomes.
Nat Commun, 13, 2022
|
|
5UP4
 
 | |
8XAT
 
 | |
8XAS
 
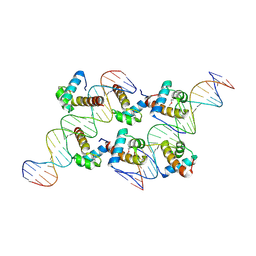 | | Crystal structure of AtARR1-DBD in complex with a DNA fragment | | Descriptor: | DNA (50-MER), Two-component response regulator ARR1 | | Authors: | Li, J.X, Zhou, C.M, zhang, P, Wang, J.W. | | Deposit date: | 2023-12-05 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6ZDJ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZET
 
 | | Crystal structure of proteinase K nanocrystals by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.701 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
6ZEV
 
 | | Crystal structure of proteinase K lamellae by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
4OV4
 
 | | Isopropylmalate synthase binding with ketoisovalerate | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, ZINC ION | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
