7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Meng, Y, Liao, H. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7EKE
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKF
 
 | | Structure of SARS-CoV-2 Alpha variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7F24
 
 | |
7F0T
 
 | |
7F23
 
 | |
7F1O
 
 | |
7F1Z
 
 | |
4Q1Q
 
 | |
3ORF
 
 | | Crystal Structure of Dihydropteridine Reductase from Dictyostelium discoideum | | Descriptor: | Dihydropteridine reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, C, Zhuang, N.N, Seo, K.H, Park, Y.S, Lee, K.H. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the dual substrate specificities of mammalian and Dictyostelium dihydropteridine reductases toward two stereoisomers of quinonoid dihydrobiopterin
Febs Lett., 585, 2011
|
|
3QNA
 
 | | Crystal structure of a 6-pyruvoyltetrahydropterin synthase homologue from Esherichia coli complexed sepiapterin | | Descriptor: | 6-carboxy-5,6,7,8-tetrahydropterin synthase, BIOPTERIN, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Lee, K.H. | | Deposit date: | 2011-02-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a novel activity of bacterial 6-pyruvoyltetrahydropterin synthase homologues distinct from mammalian 6-pyruvoyltetrahydropterin synthase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3QN9
 
 | |
3QN0
 
 | | Structure of 6-pyruvoyltetrahydropterin synthase | | Descriptor: | 6-carboxy-5,6,7,8-tetrahydropterin synthase, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Lee, K.H. | | Deposit date: | 2011-02-07 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis of a novel activity of bacterial 6-pyruvoyltetrahydropterin synthase homologues distinct from mammalian 6-pyruvoyltetrahydropterin synthase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YIU
 
 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) | | Descriptor: | Cell cycle regulatory protein GcrA | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YIW
 
 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) in complex with methylated dsDNA (crystal form 2) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, DNA (5'-D(*CP*CP*CP*TP*GP*(6MA)P*TP*TP*CP*GP*C)-3'), ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YIV
 
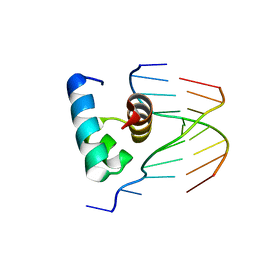 | |
5YIX
 
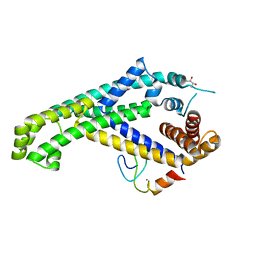 | | Caulobacter crescentus GcrA sigma-interacting domain (SID) in complex with domain 2 of sigma 70 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, RNA polymerase sigma factor RpoD, ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5ZX3
 
 | |
