8UK4
 
 | | Crystal structure of human polymerase eta with incoming dTMPnPP nucleotide opposite urea lesion | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(UD8)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Tomar, R, Egli, M, Stone, M.P. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Replication Bypass of the N -(2-Deoxy-d-erythro-pentofuranosyl)-urea DNA Lesion by Human DNA Polymerase eta.
Biochemistry, 63, 2024
|
|
8UJX
 
 | | Crystal structure of human polymerase eta with incoming dGMPnPP nucleotide opposite urea lesion | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(UD8)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Tomar, R, Egli, M, Stone, M.P. | | Deposit date: | 2023-10-11 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Replication Bypass of the N -(2-Deoxy-d-erythro-pentofuranosyl)-urea DNA Lesion by Human DNA Polymerase eta.
Biochemistry, 63, 2024
|
|
1R3G
 
 | | 1.16A X-ray structure of the synthetic DNA fragment with the incorporated 2'-O-[(2-Guanidinium)ethyl]-5-methyluridine residues | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(GMU)P*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Prakash, T.P, Puschl, A, Lesnik, E, Tereshko, V, Egli, M, Manoharan, M. | | Deposit date: | 2003-10-01 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 2'-O-[2-(Guanidinium)ethyl]-Modified Oligonucleotides: Stabilizing Effect on Duplex and Triplex Structures.
Org.Lett., 6, 2004
|
|
1NZG
 
 | | Crystal structure of A-DNA decamer GCGTA(3ME)ACGC, with a modified 5-methyluridine | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(3ME)P*AP*CP*GP*C)-3' | | Authors: | Prhavc, M, Prakash, T.P, Minasov, G, Egli, M, Manoharan, M. | | Deposit date: | 2003-02-17 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 2'-O-[2-[2-(N,N-dimethylamino)ethoxy]ethyl] modified oligonucleotides: symbiosis of charge interaction factors and stereoelectronic effects
Org.Lett., 5, 2003
|
|
1MLX
 
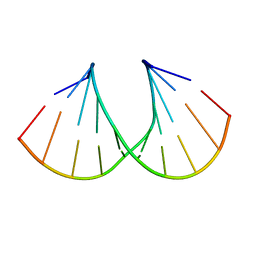 | | Crystal Structure Analysis of a 2'-O-[2-(Methylthio)-ethyl]-Modified Oligodeoxynucleotide Duplex | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*SMTP*AP*CP*GP*C)-3' | | Authors: | Prakash, T.P, Manoharan, M, Kawasaki, A.M, Fraser, A.S, Lesnik, E.A, Sioufi, N, Leeds, J.M, Teplova, M, Egli, M. | | Deposit date: | 2002-08-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 2'-O-[2-(Methylthio)ethyl]-Modified Oligonucleotide: An Analogue of 2'-O-[2-(Methoxy)-ethyl]-Modified
Oligonucleotide with Improved Protein Binding Properties and High Binding Affinity to Target RNA
Biochemistry, 41, 2002
|
|
6XHE
 
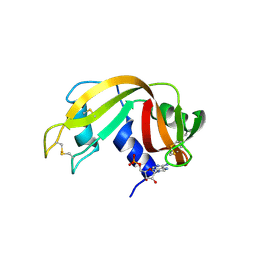 | | Structure of beta-prolinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 5'-O-[(S)-[(3S)-3-carboxypyrrolidin-1-yl](hydroxy)phosphoryl]adenosine, ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHD
 
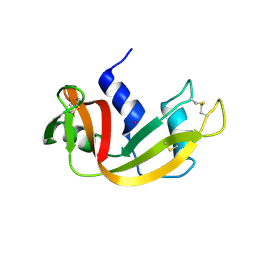 | | Structure of Prolinyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-1-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]pyrrolidine-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHF
 
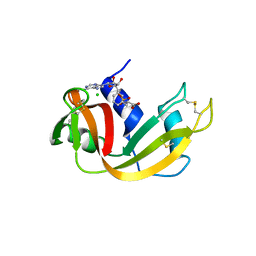 | | Structure of Phenylalanyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]-3-phenyl-propanoic acid, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHC
 
 | | Structure of glycinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]ethanoic acid, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1MA8
 
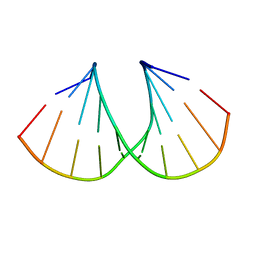 | | A-DNA decamer GCGTA(UMS)ACGC with incorporated 2'-methylseleno-uridine | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*UMSP*AP*CP*GP*C)-3' | | Authors: | Teplova, M, Wilds, C.J, Wawrzak, Z, Tereshko, V, Du, Q, Carrasco, N, Huang, Z, Egli, M. | | Deposit date: | 2002-08-01 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle
BIOCHIMIE, 84, 2002
|
|
4Y8W
 
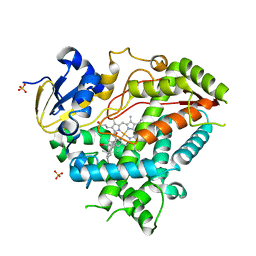 | | Crystal Structure of Human Cytochrome P450 21A2 Progesterone Complex | | Descriptor: | Cytochrome P450 21-hydroxylase, PROGESTERONE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pallan, P.S, Lei, L, Egli, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Human Cytochrome P450 21A2, the Major Steroid 21-Hydroxylase: STRUCTURE OF THE ENZYMEPROGESTERONE SUBSTRATE COMPLEX AND RATE-LIMITING C-H BOND CLEAVAGE.
J.Biol.Chem., 290, 2015
|
|
1P4F
 
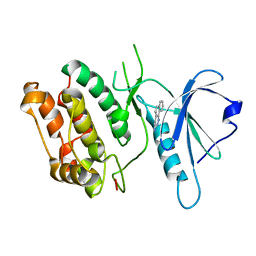 | | DEATH ASSOCIATED PROTEIN KINASE CATALYTIC DOMAIN WITH BOUND INHIBITOR FRAGMENT | | Descriptor: | 5,6-Dihydro-benzo[H]cinnolin-3-ylamine, Death-associated protein kinase 1 | | Authors: | Velentza, A.V, Wainwright, M.S, Zasadzki, M, Mirzoeva, S, Haiech, J, Focia, P.J, Egli, M, Watterson, D.M. | | Deposit date: | 2003-04-23 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An aminopyridazine-based inhibitor of a pro-apoptotic protein kinase attenuates hypoxia-ischemia induced acute brain injury.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2JEG
 
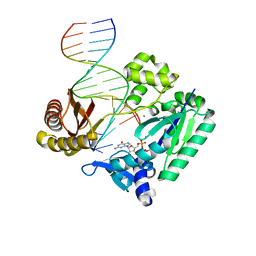 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEI
 
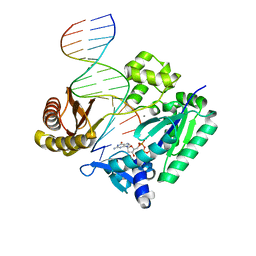 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*T)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEF
 
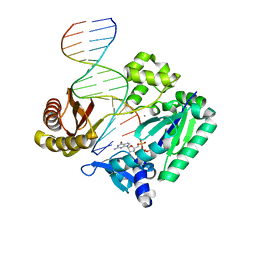 | | The Molecular Basis of Selectivity of Nucleotide Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state and X-Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*CP*BZGP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
5VR4
 
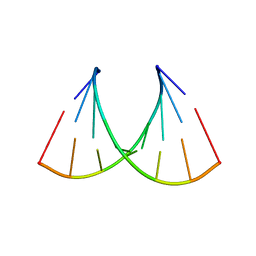 | | RNA octamer containing 2'-F-4'-OMe U. | | Descriptor: | COBALT TETRAAMMINE ION, RNA (5'-R(*CP*GP*AP*AP*(UMO)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4'-C-Methoxy-2'-deoxy-2'-fluoro Modified Ribonucleotides Improve Metabolic Stability and Elicit Efficient RNAi-Mediated Gene Silencing.
J. Am. Chem. Soc., 139, 2017
|
|
2JEJ
 
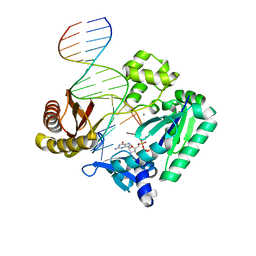 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*CP*G)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
6CY2
 
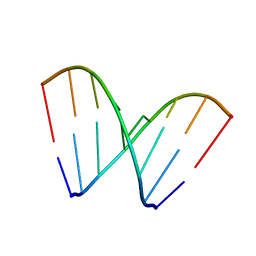 | | RNA octamer containing 2'-OMe, 4'Calpha-OMe U. | | Descriptor: | RNA (5'-R(*(CBV)P*GP*AP*AP*(UOA)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
6CXZ
 
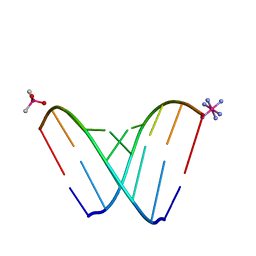 | | RNA octamer containing 2'-F, 4'-Calpha-Me U. | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*AP*AP*(U4M)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
6CY4
 
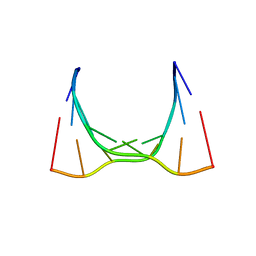 | | RNA octamer containing 2'-OMe, 4'- Cbeta-OMe U. | | Descriptor: | RNA (5'-R(*(CBV)P*GP*AP*AP*(UOB)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
5V1L
 
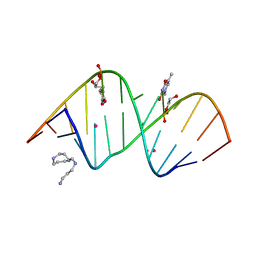 | | Structure of S-GNA dodecamer | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*(ZTH)P*AP*GP*CP*G)-3'), SPERMINE, STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
5V1K
 
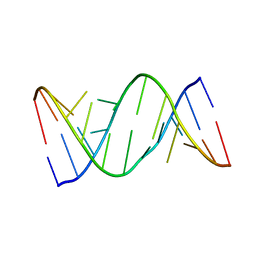 | | Structure of R-GNA dodecamer | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*(5BU)P*(8RJ)P*AP*GP*CP*G)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 2017
|
|
1RXA
 
 | |
6CY0
 
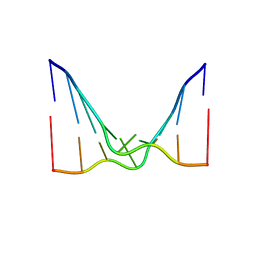 | | RNA octamer containing 2'-F, 4'-Cbeta-OMe U. | | Descriptor: | RNA (5'-R(*(CBV)P*GP*AP*AP*(UFB)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
5V2H
 
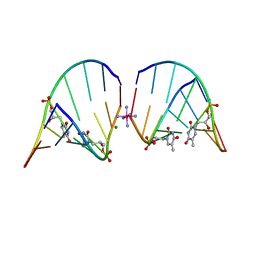 | | RNA octamer containing glycol nucleic acid, SgnT | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, RNA (5'-R(*(CBV)P*GP*AP*AP*(ZTH)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.080013 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
