2IT4
 
 | | X ray structure of the complex between Carbonic Anhydrase I and the phosphonate antiviral drug foscarnet | | Descriptor: | Carbonic anhydrase 1, PHOSPHONOFORMIC ACID, ZINC ION | | Authors: | Temperini, C, Innocenti, A, Guerri, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2006-10-19 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosph(on)ate as a zinc-binding group in metalloenzyme inhibitors: X-ray crystal structure of the antiviral drug foscarnet complexed to human carbonic anhydrase I.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6S3J
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant E134C/F149C | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Bash, Y, Rush, I, Shahar, A, Pazy, Y, Fishman, A. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridges to Stability: Engineering Disulfide Bonds Towards Enhanced Lipase Biodiesel Synthesis
Chemcatchem, 2019
|
|
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
8TG9
 
 | |
2DRE
 
 | | Crystal structure of Water-soluble chlorophyll protein from lepidium virginicum at 2.00 angstrom resolution | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Horigome, D, Satoh, H, Itoh, N, Mitsunaga, K, Oonishi, I, Nakagawa, A, Uchida, A. | | Deposit date: | 2006-06-08 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism and photoprotective function of water-soluble chlorophyll-binding protein.
J.Biol.Chem., 282, 2007
|
|
8TRC
 
 | |
8TR2
 
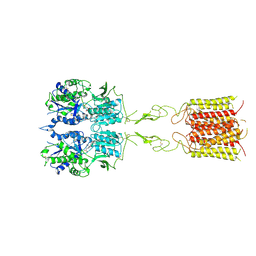 | | mGluR3 in the presence of the agonist LY379268 | | Descriptor: | (1S,4R,5R,6S)-4-amino-2-oxabicyclo[3.1.0]hexane-4,6-dicarboxylic acid, CALCIUM ION, Metabotropic glutamate receptor 3 | | Authors: | Strauss, A, Levitz, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of allosteric modulation of metabotropic glutamate receptor activation and desensitization.
Biorxiv, 2023
|
|
8TGA
 
 | |
8TQB
 
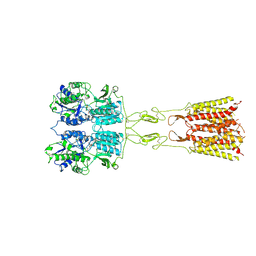 | | mGluR3 in the presence of the agonist LY379268 and PAM VU6023326 | | Descriptor: | (1R,4R,5S,6R)-4-azanyl-2-oxabicyclo[3.1.0]hexane-4,6-dicarboxylic acid, CALCIUM ION, Metabotropic glutamate receptor 3 | | Authors: | Strauss, A, Levitz, J. | | Deposit date: | 2023-08-06 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of allosteric modulation of metabotropic glutamate receptor activation and desensitization.
Biorxiv, 2023
|
|
1JB9
 
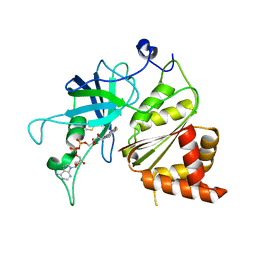 | | Crystal Structure of The Ferredoxin:NADP+ Reductase From Maize Root AT 1.7 Angstroms | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2001-06-03 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues.
Biochemistry, 40, 2001
|
|
8TR0
 
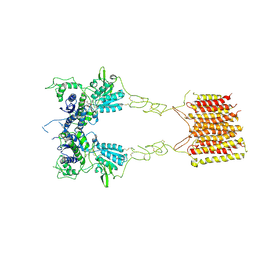 | |
6S4M
 
 | | Crystal structure of the human organic anion transporter MFSD10 (TETRAN) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Major facilitator superfamily domain-containing protein 10 | | Authors: | Pascoa, T.C, Pike, A.C.W, Bushell, S.R, Quigley, A, Chu, A, Mukhopadhyay, S.M.M, Shrestha, L, Venkaya, S, Chalk, R, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human organic anion transporter TETRAN (MFSD10)
To be published
|
|
5MJM
 
 | | Single-shot pink beam serial crystallography: Phycocyanin (Five chips merged) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5MKG
 
 | | PA3825-EAL Ca-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Diguanylate phosphodiesterase | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
8PQN
 
 | | NQO1 bound to RBS-10 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1, ~{N}-[4-[(3-methylphenyl)carbonylamino]phenyl]-5-nitro-furan-2-carboxamide | | Authors: | Pous, J, Jose-Duran, F, Mayor-Ruiz, C, Riera, A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Discovery and Mechanistic Elucidation of NQO1-Bioactivatable Small Molecules That Overcome Resistance to Degraders.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1JIQ
 
 | | Crystal Structure of Human Autocrine Motility Factor | | Descriptor: | autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
8PVV
 
 | | Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 30 nt RNA guide and 51 nt DNA target | | Descriptor: | Archaeoglobus fulgidus AfAgo-N protein, DNA (51-MER), MAGNESIUM ION, ... | | Authors: | Manakova, E.N, Zaremba, M, Pocevicuite, R, Golovinas, E, Sasnauskas, G, Zagorskaite, E, Silanskas, A. | | Deposit date: | 2023-07-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
8Q0R
 
 | | X-ray structure of MNEI mutant Mut9 (E23A, C41A, Y65R, S76Y) | | Descriptor: | ACETATE ION, Monellin chain B,Monellin chain A, SULFATE ION | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
1JQH
 
 | | IGF-1 receptor kinase domain | | Descriptor: | IGF-1 receptor kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pautsch, A, Zoephel, A, Ahorn, H, Spevak, W, Hauptmann, R, Nar, H. | | Deposit date: | 2001-08-07 | | Release date: | 2002-04-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bisphosphorylated IGF-1 receptor kinase: insight into domain movements upon kinase activation.
Structure, 9, 2001
|
|
1U0D
 
 | | Y33H Mutant of Homing endonuclease I-CreI | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', DNA endonuclease I-CreI | | Authors: | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
8PLZ
 
 | | Cryo-EM structure of CAK in complex with inhibitor CT7030 | | Descriptor: | (3~{R},4~{R})-4-[[[7-[(2-methoxyphenyl)methylamino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-06-27 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
1UF2
 
 | | The Atomic Structure of Rice dwarf Virus (RDV) | | Descriptor: | Core protein P3, Outer capsid protein P8, Structural protein P7 | | Authors: | Nakagawa, A, Miyazaki, N, Taka, J, Naitow, H, Ogawa, A, Fujimoto, Z, Mizuno, H, Higashi, T, Watanabe, Y, Omura, T, Cheng, R.H, Tsukihara, T. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Structure, 11, 2003
|
|
1U4L
 
 | | human RANTES complexed to heparin-derived disaccharide I-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
2JE5
 
 | | STRUCTURAL AND MECHANISTIC BASIS OF PENICILLIN BINDING PROTEIN INHIBITION BY LACTIVICINS | | Descriptor: | (2E)-2-{[(2S)-2-(ACETYLAMINO)-2-CARBOXYETHOXY]IMINO}PENTANEDIOIC ACID, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
8U3M
 
 | | The FARFAR-MD-NMR ensemble of an HIV-1 TAR excited state | | Descriptor: | The excited state of HIV-1 transactivation response element (31-MER) | | Authors: | Geng, A, Ganser, L, Roy, R, Shi, H, Pratihar, S, Case, D.A, Al-Hashimi, H.M. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA excited conformational state at atomic resolution.
Nat Commun, 14, 2023
|
|
