4XYF
 
 | | Crystal structure of c-Met in complex with (S)-5-(8-fluoro-3-(1-(3-(2-methoxyethoxy)quinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1S)-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-(2-methoxyethoxy)quinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
8HP3
 
 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HP0
 
 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, SULFATE ION | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HPE
 
 | | Crystal structure of Leucine dehydrogenase | | Descriptor: | GLYCEROL, Leucine dehydrogenase, SULFATE ION | | Authors: | Li, X, Song, W. | | Deposit date: | 2022-12-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Chembiochem, 24, 2023
|
|
7UM7
 
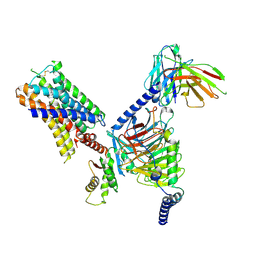 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Methylergometrine | | Descriptor: | (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM5
 
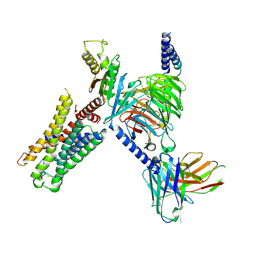 | | CryoEM structure of Go-coupled 5-HT5AR in complex with 5-CT | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM4
 
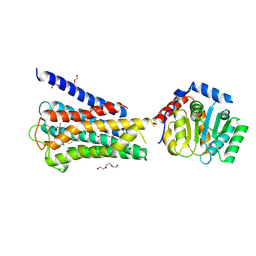 | | Crystal structure of inactive 5-HT5AR in complex with AS2674723 | | Descriptor: | 5-hydroxytryptamine receptor 5A, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Zhang, S, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM6
 
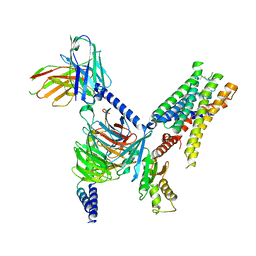 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride | | Descriptor: | 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V69
 
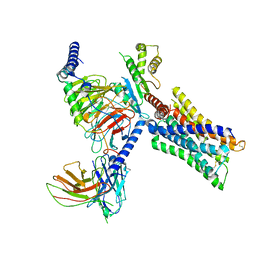 | | Cryo-EM structure of a class A GPCR-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V6A
 
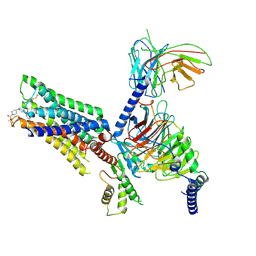 | | Cry-EM structure of M4-c110-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V68
 
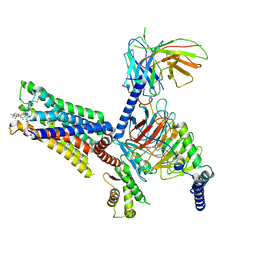 | | An Agonist and PAM-bound Class A GPCR with Gi protein complex structure | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, J.J, Wu, L.J, Wu, M, Hua, T, Liu, Z.J, Wang, T. | | Deposit date: | 2021-08-20 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
8OWG
 
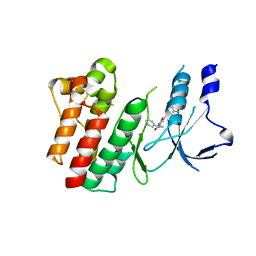 | | Crystal structure of D1228V c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUU
 
 | | Crystal structure of D1228V c-MET bound by compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3-ethynyl-5-fluoranyl-1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, FORMIC ACID, ... | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OVZ
 
 | | Crystal structure of D1228V c-MET bound by compound 16 | | Descriptor: | 1-[(1S)-1-[3-(1H-imidazol-4-yl)phenyl]ethyl]-5-(1H-indazol-7-yl)pyrimidine-2,4-dione, Hepatocyte growth factor receptor, IODIDE ION | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8HR6
 
 | |
8OUV
 
 | | Crystal structure of D1228V c-MET bound by compound 15 | | Descriptor: | 5-(1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OW3
 
 | | Crystal structure of wild-type c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OV7
 
 | | Crystal structure of D1228V c-MET bound by compound 10 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-[3-(1H-imidazol-5-yl)phenyl]ethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
5EXH
 
 | |
2ZIH
 
 | |
1RTW
 
 | | X-ray Structure of PF1337, a TenA Homologue from Pyrococcus furiosus. Northeast Structural Genomics Research Consortium (Nesg) Target PFR34 | | Descriptor: | (4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL DIHYDROGEN PHOSPHATE, PHOSPHATE ION, transcriptional activator, ... | | Authors: | Benach, J, Edstrom, W.C, Lee, I, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The 2.35 A structure of the TenA homolog from Pyrococcus furiosus supports an enzymatic function in thiamine metabolism.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2KCV
 
 | | Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c | | Descriptor: | Tetratricopeptide repeat domain protein | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c
To be Published
|
|
4HXX
 
 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | Descriptor: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
6LS5
 
 | | Structure of human liver FBPase complexed with covalent allosteric inhibitor | | Descriptor: | 2-(ethyldisulfanyl)-1,3-benzothiazole, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Yunyuan, H, Rongrong, S, Yixiang, X, Shuaishuai, N, Yanliang, R, Jian, L, Jian, W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Identification of the New Covalent Allosteric Binding Site of Fructose-1,6-bisphosphatase with Disulfiram Derivatives toward Glucose Reduction.
J.Med.Chem., 63, 2020
|
|
