7Y1J
 
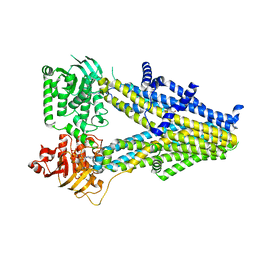 | | Structure of SUR2A in complex with Mg-ATP and repaglinide in the inward-facing conformation. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 9, MAGNESIUM ION, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1L
 
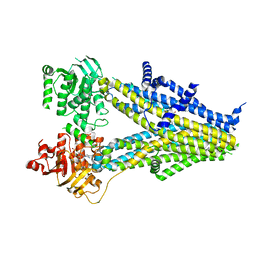 | | Structure of SUR2B in complex with Mg-ATP and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, MAGNESIUM ION, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1K
 
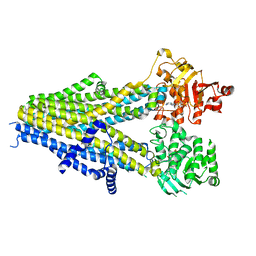 | | Structure of SUR2A in complex with Mg-ATP, Mg-ADP and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1M
 
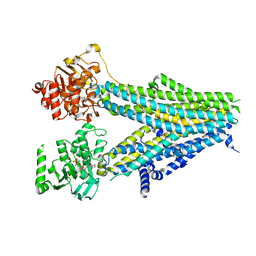 | | Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1N
 
 | | Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the partially occluded state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7YFK
 
 | | The structure of human pregnane X receptor in complex with an SRC-1 coactivator peptide and a limonoid compound, nomilin | | Descriptor: | Nomilin, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 | | Authors: | Xia, Y, Yao, D, Huang, C, Cao, Y. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pregnane X receptor agonist nomilin extends lifespan and healthspan in preclinical models through detoxification functions.
Nat Commun, 14, 2023
|
|
2GNI
 
 | | PKA fivefold mutant model of Rho-kinase with inhibitor Fasudil (HA1077) | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, cAMP-dependent protein kinase inhibitor alpha, cAMP-dependent protein kinase, ... | | Authors: | Bonn, S, Herrero, S, Breitenlechner, C.B, Engh, R.A, Gassel, M, Bossemeyer, D. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity
J.Biol.Chem., 281, 2006
|
|
3T59
 
 | |
5RL2
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-26 (Mpro-x3115) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(2-methoxyethyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5UNO
 
 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5RL0
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-2 (Mpro-x3110) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ethyl N-[(2R)-2-[(4-tert-butylphenyl)(propanoyl)amino]-2-(pyridin-3-yl)acetyl]-beta-alaninate | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
3QLB
 
 | | Enantiopyochelin outer membrane TonB-dependent transporter from Pseudomonas fluorescens bound to the ferri-enantiopyochelin | | Descriptor: | CITRIC ACID, ENANTIO-PYOCHELIN FE(III), Enantio-pyochelin receptor, ... | | Authors: | Brillet, K, Noel, S, Mislin, G.L.A, Reimmann, C, Schalk, I.J, Cobessi, D. | | Deposit date: | 2011-02-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Pyochelin enantiomers and their outer-membrane siderophore transporters in fluorescent pseudomonads: structural bases for unique enantiospecific recognition
J.Am.Chem.Soc., 133, 2011
|
|
2HLP
 
 | | CRYSTAL STRUCTURE OF THE E267R MUTANT OF A HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
2HOX
 
 | | alliinase from allium sativum (garlic) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimon, L.J.W, Rabinkov, A, Wilcheck, M, Mirelman, D, Frolow, F. | | Deposit date: | 2006-07-17 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two Structures of Alliinase from Alliium sativum L.: Apo Form and Ternary Complex with Aminoacrylate Reaction Intermediate Covalently Bound to the PLP Cofactor.
J.Mol.Biol., 366, 2007
|
|
2KE8
 
 | | NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Johannsen, S, Duepre, N, Boehme, D, Mueller, J, Sigel, R.K.O. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA double helix with consecutive metal-mediated base pairs.
Nat.Chem., 2, 2010
|
|
3UAK
 
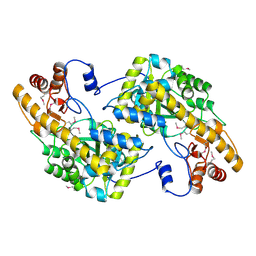 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | Descriptor: | De Novo designed cysteine esterase ECH14 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
2HSP
 
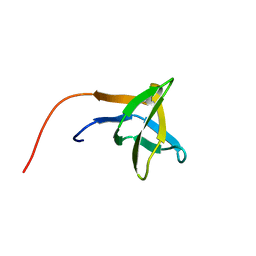 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
2I1Y
 
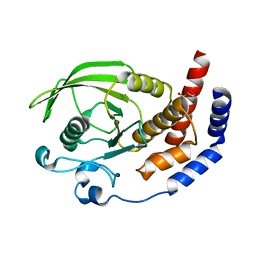 | | Crystal structure of the phosphatase domain of human PTP IA-2 | | Descriptor: | GLYCEROL, Receptor-type tyrosine-protein phosphatase | | Authors: | Faber-Barata, J, Patskovsky, Y, Alvarado, J, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Maletic, M, Rooney, I, Bain, K.T, Freeman, M, Russell, J.C, Thompson, D.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2HY3
 
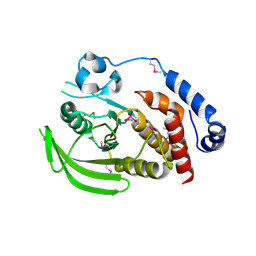 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2KIZ
 
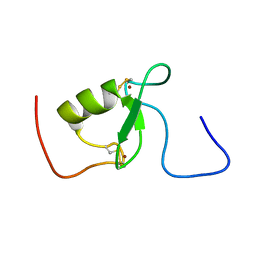 | | Solution structure of Arkadia RING-H2 finger domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Kandias, N.G, Chasapis, C.T, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2013-03-20 | | Method: | SOLUTION NMR | | Cite: | NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase.
Proteins, 80, 2012
|
|
7AAD
 
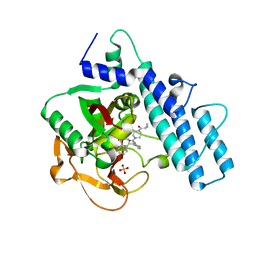 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
8OQG
 
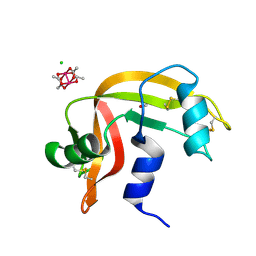 | | Cross-linked crystal of dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (high temperature data collection) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8P4Z
 
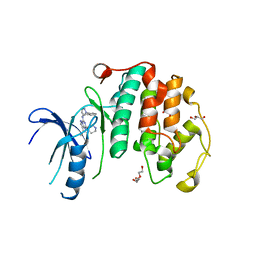 | | Crystal structure of the human CDK7 kinase domain in complex with LDC4297 | | Descriptor: | 2-[(3R)-piperidin-3-yl]oxy-8-propan-2-yl-N-[(2-pyrazol-1-ylphenyl)methyl]pyrazolo[1,5-a][1,3,5]triazin-4-amine, Cyclin-dependent kinase 7, GLYCEROL, ... | | Authors: | Laursen, M, Caing-Carlsson, R, Houssari, R, Javadi, A, Kimbung, Y.R, Murina, V, Orozco-Rodriguez, J.M, Svensson, A, Welin, M, Logan, D, Svensson, B, Walse, B. | | Deposit date: | 2023-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the human CDK7 kinase domain in complex with LDC4297
To Be Published
|
|
8OWA
 
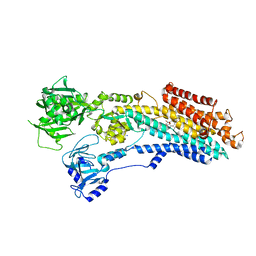 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-4 | | Descriptor: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | Authors: | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
8OQD
 
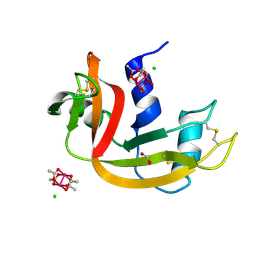 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (1 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
