7EW6
 
 | | Barley photosystem I-LHCI-Lhca5 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-24 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
3T35
 
 | | Arabidopsis thaliana dynamin-related protein 1A in postfission state | | Descriptor: | Dynamin-related protein 1A, LINKER, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yan, L.M, Ma, Y.Y, Sun, Y.N, Lou, Z.Y. | | Deposit date: | 2011-07-24 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.592 Å) | | Cite: | Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
J Mol Cell Biol, 3, 2011
|
|
4ESS
 
 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
7WGX
 
 | | SARS-CoV-2 spike glycoprotein trimer in closed state after treatment with Cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGY
 
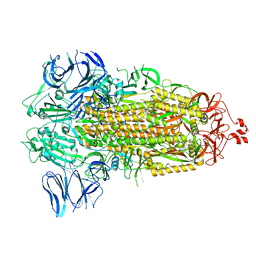 | | SARS-CoV-2 spike glycoprotein trimer in Intermediate state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGZ
 
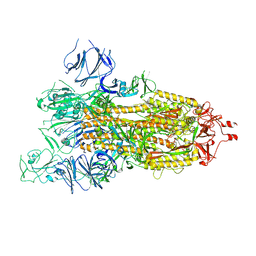 | | SARS-CoV-2 spike glycoprotein trimer in open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGV
 
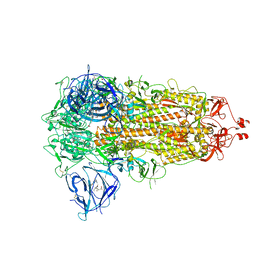 | | SARS-CoV-2 spike glycoprotein trimer in closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7EU3
 
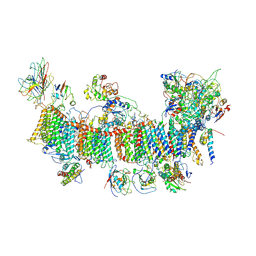 | | Chloroplast NDH complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EWK
 
 | | Barley photosystem I-LHCI-Lhca6 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3OF6
 
 | | Human pre-T cell receptor crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre T-cell antigen receptor alpha, T cell receptor beta chain | | Authors: | Pang, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Nature, 467, 2010
|
|
4F2V
 
 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3TY0
 
 | | Structure of PPARgamma ligand binding domain in complex with (R)-5-(3-((3-(6-methoxybenzo[d]isoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)methyl)phenyl)-5-methyloxazolidine-2,4-dione | | Descriptor: | (5R)-5-(3-{[3-(6-methoxy-1,2-benzoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl]methyl}phenyl)-5-methyl-1,3-oxazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Soisson, S.M, Meinke, P.M, McKeever, B, Liu, W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Benzimidazolones: a new class of selective peroxisome proliferator-activated receptor gamma (PPAR-gamma) modulators.
J.Med.Chem., 54, 2011
|
|
3TP4
 
 | | Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3U3E
 
 | |
6HL9
 
 | |
2LCJ
 
 | |
2M77
 
 | | [Asp2]RTD-1 | | Descriptor: | [Asp2]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
2M78
 
 | | [Asp11]RTD-1 | | Descriptor: | [Asp11]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
2M79
 
 | | [Asp2,11]RTD-1 | | Descriptor: | [Asp2,11]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
6GPE
 
 | |
6GPN
 
 | |
2INW
 
 | | Crystal structure of Q83JN9 from Shigella flexneri at high resolution. Northeast Structural Genomics Consortium target SfR137. | | Descriptor: | PHOSPHATE ION, Putative structural protein | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Vorobiev, S.M, Wang, D, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-09 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
6HL8
 
 | |
