4H7E
 
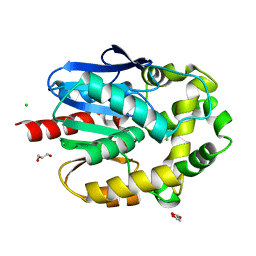 | | Crystal structure of haloalkane dehalogenase LinB V112A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7F
 
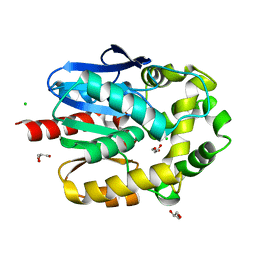 | | Crystal structure of haloalkane dehalogenase LinB V134I mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4GS3
 
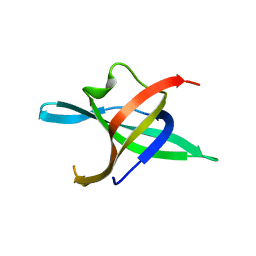 | | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tencongensis solved ab initio | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Liebschner, D, Brzezinski, K, Dauter, M, Dauter, Z, Nowak, M, Kur, J, Olszewski, M. | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tengcongensis solved ab initio.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7X73
 
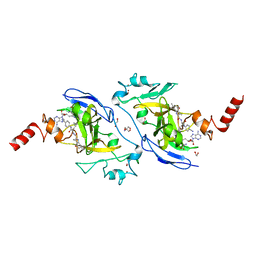 | | Structure of G9a in complex with RK-701 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nat Commun, 14, 2023
|
|
4HD4
 
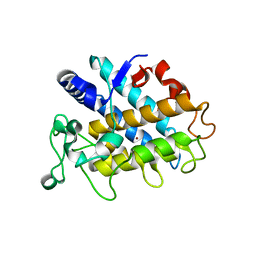 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218F mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4HD7
 
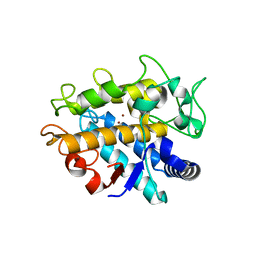 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218G mutant soaked in CuSO4 | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4HGH
 
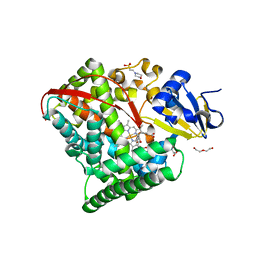 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset I) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
5LS0
 
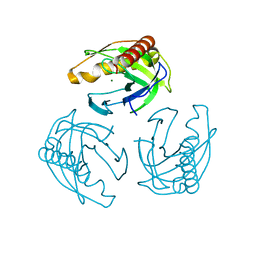 | | Crystal structure of Inorganic Pyrophosphatase PPA1 from Arabidopsis thaliana | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Soluble inorganic pyrophosphatase 1 | | Authors: | Grzechowiak, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of plant inorganic pyrophosphatase, an enzyme with a moonlighting autoproteolytic activity.
Biochem.J., 476, 2019
|
|
4HGF
 
 | | Crystal structure of P450 BM3 5F5K heme domain variant complexed with styrene | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4U3U
 
 | | Crystal structure of Cycloheximide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4UFS
 
 | |
4U5U
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[3-(pyridin-3-yl)benzyl]-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U6F
 
 | | Crystal structure of T-2 toxin bound to the yeast 80S ribosome | | Descriptor: | 12,13-Epoxytrichothec-9-ene-3,4,8,15-tetrol-4,15-diacetate-8-isovalerate, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-28 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U1V
 
 | | Crystal structure of the E. coli ribosome bound to linopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4U3N
 
 | | Crystal structure of CCA trinucleotide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U58
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzoyl]-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5O
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-D-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U4Z
 
 | | Crystal structure of Phyllanthoside bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
5WBN
 
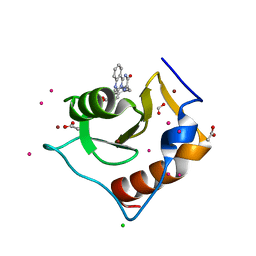 | | Crystal structure of fragment 3-(3-Benzyl-2-oxo-2H-[1,2,4]triazino[2,3-c]quinazolin-6-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-benzyl-2-oxo-2H-[1,2,4]triazino[2,3-c]quinazolin-6-yl)propanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Harding, R.J, Walker, J.R, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of fragment 3-(3-Benzyl-2-oxo-2H-[1,2,4]triazino[2,3-c]quinazolin-6-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain
To be published
|
|
4UAR
 
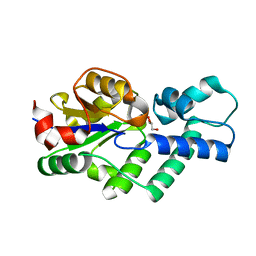 | | Crystal structure of apo-CbbY from Rhodobacter sphaeroides | | Descriptor: | GLYCEROL, Protein CbbY | | Authors: | Bracher, A, Sharma, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2014-08-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degradation of potent Rubisco inhibitor by selective sugar phosphatase.
Nat.Plants, 1, 2015
|
|
4UAQ
 
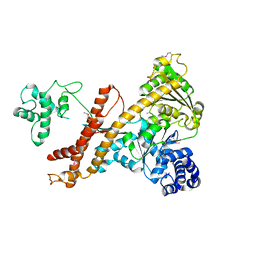 | | Crystal structure of the accessory translocation ATPase, SecA2, from Mycobacterium tuberculosis | | Descriptor: | Protein translocase subunit SecA 2 | | Authors: | Swanson-Smith, S, Ioerger, T.R, Rigel, N.W, Miller, B.K, Braunstein, M, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-09 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Similarities and Differences between Two Functionally Distinct SecA Proteins, Mycobacterium tuberculosis SecA1 and SecA2.
J.Bacteriol., 198, 2015
|
|
4UFT
 
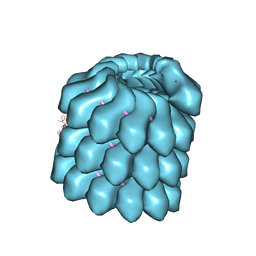 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
4UFR
 
 | |
4U4T
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound inward-open state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4U
 
 | | Crystal structure of Lycorine bound to the yeast 80S ribosome | | Descriptor: | (1S,2S,12bS,12cS)-2,4,5,7,12b,12c-hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
