7OT9
 
 | |
6HRC
 
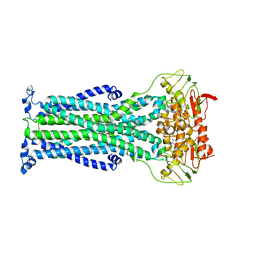 | | Outward-facing PglK with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, WlaB protein | | Authors: | Perez, C, Locher, K.P. | | Deposit date: | 2018-09-26 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Outward-Facing PglK and Molecular Dynamics of Lipid-Linked Oligosaccharide Recognition and Translocation.
Structure, 27, 2019
|
|
6HFB
 
 | | Outward-facing conformation of a multidrug resistance MATE family transporter of the MOP superfamily. | | Descriptor: | CESIUM ION, Uncharacterized protein | | Authors: | Nonaka, T, Zakrzewska, S, Safarian, S, Michel, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6YK8
 
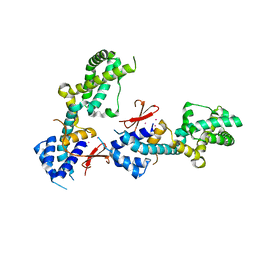 | | OTU-like deubiquitinase from Legionella -Lpg2529 | | Descriptor: | PLATINUM (II) ION, Uncharacterized protein | | Authors: | Shin, D, Dikic, I. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Bacterial OTU deubiquitinases regulate substrate ubiquitination upon Legionella infection.
Elife, 9, 2020
|
|
7PHR
 
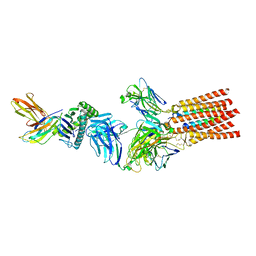 | | Structure of a fully assembled T-cell receptor engaging a tumor-associated peptide-MHC I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Susac, L, Thomas, C, Tampe, R. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC.
Cell, 185, 2022
|
|
6VK3
 
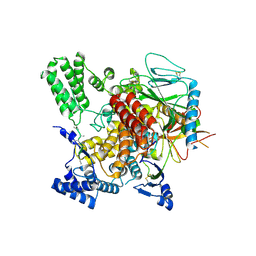 | | CryoEM structure of Hrd3/Yos9 complex | | Descriptor: | Hrd3, Protein OS-9 homolog | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VJZ
 
 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 complex of the expected topology | | Descriptor: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VJY
 
 | | Cryo-EM structure of Hrd1/Hrd3 monomer | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
5MKK
 
 | | Crystal structure of the heterodimeric ABC transporter TmrAB, a homolog of the antigen translocation complex TAP | | Descriptor: | Multidrug resistance ABC transporter ATP-binding and permease protein, SULFATE ION | | Authors: | Noell, A, Thomas, C, Tomasiak, T.M, Olieric, V, Wang, M, Diederichs, K, Stroud, R.M, Pos, K.M, Tampe, R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7R1Y
 
 | |
6VK1
 
 | | CryoEM structure of Hrd1/Hrd3 part from Hrd1-Usa1/Der1/Hrd3 complex | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
6VK0
 
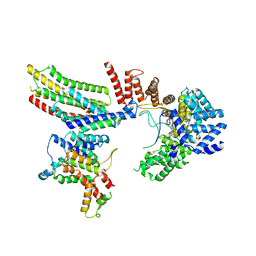 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 of the flipped topology | | Descriptor: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
2B6X
 
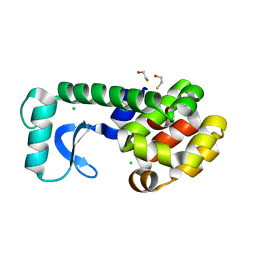 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6Y
 
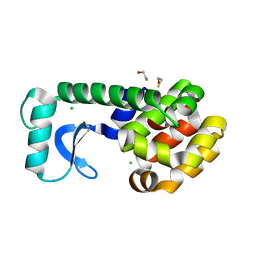 | | T4 Lysozyme mutant L99A at ambient pressure | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6T
 
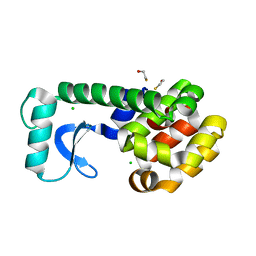 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural rigidity of a large cavity-containing protein revealed by high-pressure crystallography.
J.Mol.Biol., 367, 2007
|
|
2B74
 
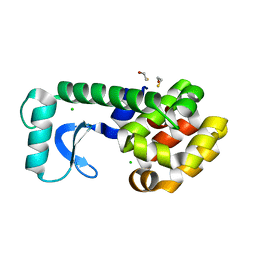 | | T4 Lysozyme mutant L99A at 100 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B73
 
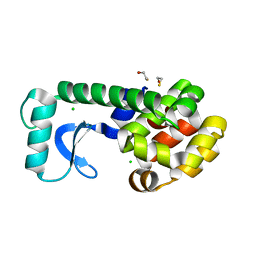 | | T4 Lysozyme mutant L99A at 100 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4MLB
 
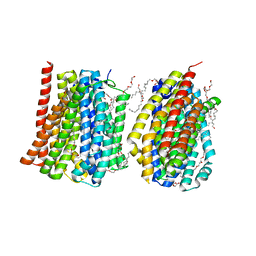 | | Reverse polarity of binding pocket suggests different function of a MOP superfamily transporter from Pyrococcus furiosus Vc1 (DSM3638) | | Descriptor: | CHLORIDE ION, PENTAETHYLENE GLYCOL MONODECYL ETHER, PF0708 | | Authors: | Malviya, V.N, Nonaka, T, Muenke, C, Koepke, J, Michel, H. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2OE4
 
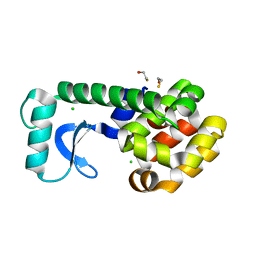 | | High Pressure Psuedo Wild Type T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a non-polar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8PEE
 
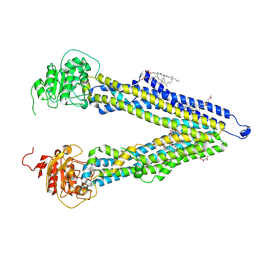 | | ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
6RU6
 
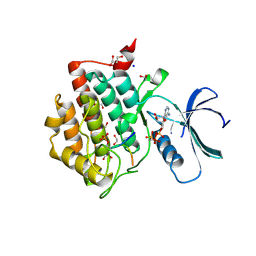 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with monophosphorylated p63 PAD1P peptide | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6RYB
 
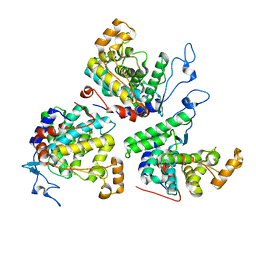 | |
6RU7
 
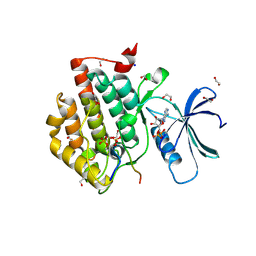 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with double phosphorylated p63 PAD2P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6FHZ
 
 | |
6RYA
 
 | | Structure of Dup1 mutant H67A:Ubiquitin complex | | Descriptor: | Polyubiquitin-C, Septation initiation protein | | Authors: | Donghyuk, S, Ivan, D. | | Deposit date: | 2019-06-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB.
Mol.Cell, 77, 2020
|
|
