6WXN
 
 | | EGFR(T790M/V948R) in complex with LN3844 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
6XL4
 
 | | EGFR(T790M/V948R) in complex with AZD9291 and DDC4002 | | Descriptor: | 10-benzyl-8-fluoro-5,10-dihydro-11H-dibenzo[b,e][1,4]diazepin-11-one, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors.
Nat Commun, 13, 2022
|
|
2F31
 
 | |
3IKA
 
 | |
2ITO
 
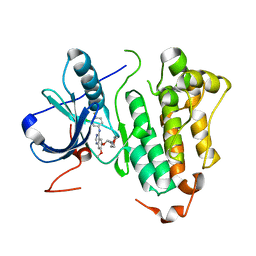 | | Crystal structure of EGFR kinase domain G719S mutation in complex with Iressa | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, Gefitinib | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2IUH
 
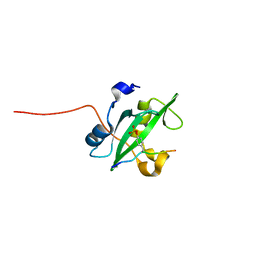 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with c-Kit phosphotyrosyl peptide | | Descriptor: | C-KIT PHOSPHOTYROSYL PEPTIDE, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUG
 
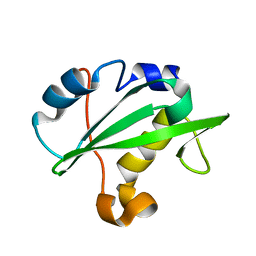 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUI
 
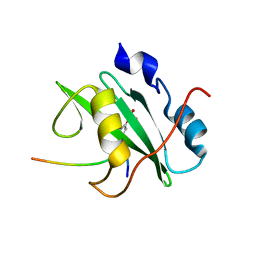 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with PDGFR phosphotyrosyl peptide | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Platelet-derived growth factor receptor beta | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino- Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2J5E
 
 | |
2ITU
 
 | | Crystal structure of EGFR kinase domain L858R mutation in complex with AFN941 | | Descriptor: | 1,2,3,4-Tetrahydrogen Staurosporine, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
5TX3
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-[(1S)-4-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-2-({3-[(pyrrolidin-1-yl)methyl]phenyl}amino)-5,7-dihydro-6H-pyrrolo[2,3-d]pyrimidin-6-one, Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
4IM2
 
 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | CHLORIDE ION, N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IM0
 
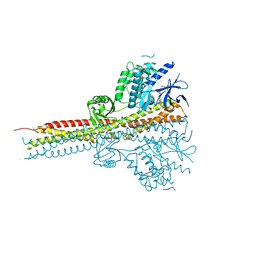 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(morpholin-4-ylmethyl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4001 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IM3
 
 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, ... | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.342 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
5WIN
 
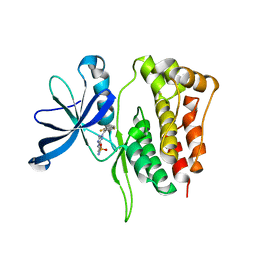 | | JAK2 Pseudokinase in complex with JNJ7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5TWU
 
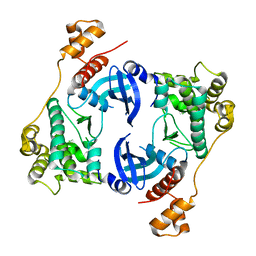 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5WFN
 
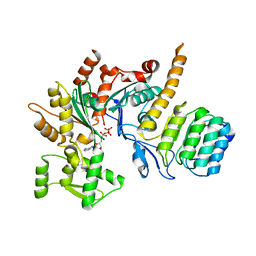 | | Revised model of leiomodin 2-mediated actin regulation (alternate refinement of PDB 4RWT) | | Descriptor: | Actin-5C, Leiomodin-2, MAGNESIUM ION, ... | | Authors: | Yurtsever, Z, Eck, M.J, Dominguez, R. | | Deposit date: | 2017-07-12 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of leiomodin 2 in complex with actin: a structural and functional reexamination
Biophys.J., 113, 2017
|
|
5WIJ
 
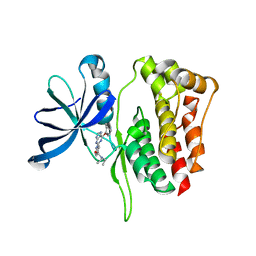 | | JAK2 Pseudokinase in complex with NU6140 | | Descriptor: | 4-{[6-(cyclohexylmethoxy)-7H-purin-2-yl]amino}-N,N-diethylbenzamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIL
 
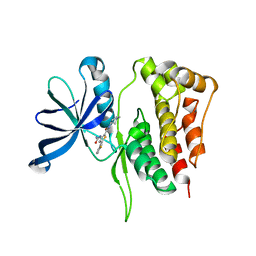 | | JAK2 Pseudokinase in complex with AZD7762 | | Descriptor: | 5-(3-fluorophenyl)-N-[(3S)-3-piperidyl]-3-ureido-thiophene-2-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIK
 
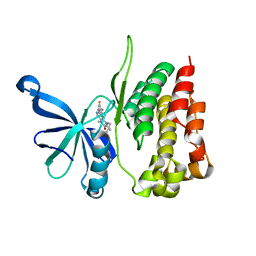 | | JAK2 Pseudokinase in complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIM
 
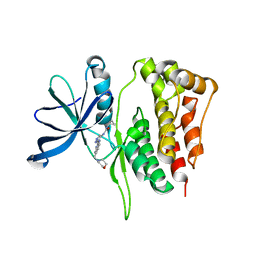 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
4L00
 
 | |
4L01
 
 | |
4LL0
 
 | | EGFR L858R/T790M in complex with PD168393 | | Descriptor: | Epidermal growth factor receptor, N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}propanamide | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Mechanism for activation of mutated epidermal growth factor receptors in lung cancer.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7KXZ
 
 | | Active conformation of EGFR kinase in complex with BI-4020 | | Descriptor: | (20R)-10,15,20-trimethyl-2-[(4-methylpiperazin-1-yl)methyl]-18,19,20,21-tetrahydro-15H,17H-12,8-(metheno)pyrazolo[3',4':2,3][1,5,10,12]oxatriazacycloheptadecino[12,11-a]benzimidazol-7(6H)-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-12-06 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of the Macrocyclic Inhibitor BI-4020 Binding to EGFR Kinase.
Chemmedchem, 2024
|
|
