4FR3
 
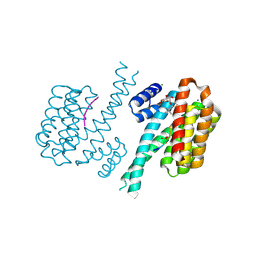 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide and stabilizer 16-O-Me-FC-H | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, MAGNESIUM ION, ... | | Authors: | Ottmann, C, Anders, C, Schumacher, B. | | Deposit date: | 2012-06-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of k(+) channels at the cell surface.
Chem.Biol., 20, 2013
|
|
4FRL
 
 | | Crystal Structure of BBBB+UDP+Gal at pH 8.0 with MPD as the cryoprotectant | | Descriptor: | Histo-blood group ABO system transferase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
4MBX
 
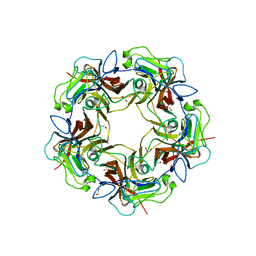 | | Structure of unliganded B-Lymphotropic Polyomavirus VP1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Major Capsid Protein VP1 | | Authors: | Khan, Z.M, Neu, U, Schuch, B, Stehle, T. | | Deposit date: | 2013-08-21 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of B-Lymphotropic Polyomavirus VP1 in Complex with Oligosaccharide Ligands.
Plos Pathog., 9, 2013
|
|
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | COLD SHOCK PROTEIN TMCSP | | Authors: | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | Deposit date: | 2000-11-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
3O5C
 
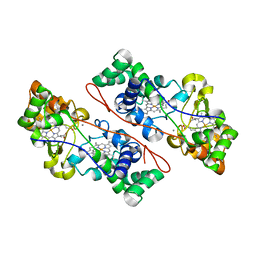 | | Cytochrome c Peroxidase BccP of Shewanella oneidensis | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, Cytochrome c551 peroxidase, ... | | Authors: | Seidel, J, Einsle, O. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the Electron Transport Chain to and the Catalytic Activity of the Diheme Cytochrome c Peroxidase CcpA of Shewanella oneidensis.
Appl.Environ.Microbiol., 77, 2011
|
|
7YTL
 
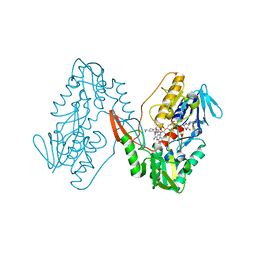 | | Structure of a oxidoreductase in complex with quinone | | Descriptor: | Apoptosis inducing factor mitochondria associated 2, FLAVIN-ADENINE DINUCLEOTIDE, UBIQUINONE-1 | | Authors: | Lv, Y, Sun, Q, Wang, Q, Zhu, D. | | Deposit date: | 2022-08-15 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into FSP1 catalysis and ferroptosis inhibition.
Nat Commun, 14, 2023
|
|
7XPI
 
 | | Structure of a oxidoreductase | | Descriptor: | Apoptosis inducing factor mitochondria associated 2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Lv, Y, Sun, Q, Wang, Q, Zhu, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into FSP1 catalysis and ferroptosis inhibition.
Nat Commun, 14, 2023
|
|
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8VB1
 
 | | Crystal structure of HIV-1 protease with GS-9770 | | Descriptor: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | Authors: | Lansdon, E.B. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
8DV6
 
 | | Zika virus envelope protein structure in complex with a potent Human mAb | | Descriptor: | Envelope protein E, mAb Fab Heavy Chain, mAb Fab Light Chain | | Authors: | Cameron, A, Puhl, A.C, deSilva, A.M, Premkumar, L. | | Deposit date: | 2022-07-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure and neutralization mechanism of a human antibody targeting a complex Epitope on Zika virus.
Plos Pathog., 19, 2023
|
|
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
7YAR
 
 | | ZIKV_Fab_G9E | | Descriptor: | E protein, M protein, heavy chain, ... | | Authors: | Shu, B, Thiam-Seng, N, Lok, S. | | Deposit date: | 2022-06-28 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structure and neutralization mechanism of a human antibody targeting a complex Epitope on Zika virus.
Plos Pathog., 19, 2023
|
|
8A7I
 
 | | Crystal structure of BRD9 bromodomain in complex with compound EA-89 | | Descriptor: | Bromodomain-containing protein 9, ~{N}-[1,1-bis(oxidanylidene)thian-4-yl]-7-[3-methyl-1-(piperidin-4-ylmethyl)indol-5-yl]-4-oxidanylidene-5-propyl-thieno[3,2-c]pyridine-2-carboxamide | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | BRD9 degraders as chemosensitizers in acute leukemia and multiple myeloma.
Blood Cancer J, 12, 2022
|
|
5KRE
 
 | | Covalent inhibitor of LYPLAL1 | | Descriptor: | (2~{R})-2-phenylpiperidine-1-carbaldehyde, Lysophospholipase-like protein 1, NITRATE ION | | Authors: | Pandit, J. | | Deposit date: | 2016-07-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Selective Covalent Inhibitor of Lysophospholipase-like 1 (LYPLAL1) as a Tool to Evaluate the Role of this Serine Hydrolase in Metabolism.
Acs Chem.Biol., 11, 2016
|
|
6BJM
 
 | | Human ABO(H) blood group glycosyltransferase GTB R188K mutant | | Descriptor: | ABO blood group (Transferase A, alpha 1-3-N-acetylgalactosaminyltransferase transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Gagnon, S.M.L, Legg, M.S.G, Evans, S.V. | | Deposit date: | 2017-11-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conserved residues Arg188 and Asp302 are critical for active site organization and catalysis in human ABO(H) blood group A and B glycosyltransferases.
Glycobiology, 28, 2018
|
|
6BJJ
 
 | | Human ABO(H) blood group glycosyltransferase GTB D302A mutant | | Descriptor: | ABO blood group (Transferase A, alpha 1-3-N-acetylgalactosaminyltransferase transferase B, alpha 1-3-galactosyltransferase) | | Authors: | Gagnon, S.M.L, Legg, M.S.G, Evans, S.V. | | Deposit date: | 2017-11-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conserved residues Arg188 and Asp302 are critical for active site organization and catalysis in human ABO(H) blood group A and B glycosyltransferases.
Glycobiology, 28, 2018
|
|
6BJI
 
 | |
6BJL
 
 | | Human ABO(H) blood group glycosyltransferase GTB D302L mutant | | Descriptor: | ABO blood group (Transferase A, alpha 1-3-N-acetylgalactosaminyltransferase transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Gagnon, S.M.L, Legg, M.S.G, Evans, S.V. | | Deposit date: | 2017-11-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Conserved residues Arg188 and Asp302 are critical for active site organization and catalysis in human ABO(H) blood group A and B glycosyltransferases.
Glycobiology, 28, 2018
|
|
6BJK
 
 | | Human ABO(H) blood group glycosyltransferase GTB D302C mutant | | Descriptor: | ABO blood group (Transferase A, alpha 1-3-N-acetylgalactosaminyltransferase transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Gagnon, S.M.L, Legg, M.S.G, Evans, S.V. | | Deposit date: | 2017-11-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Conserved residues Arg188 and Asp302 are critical for active site organization and catalysis in human ABO(H) blood group A and B glycosyltransferases.
Glycobiology, 28, 2018
|
|
7L5D
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with demethylated analog of masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
5KHU
 
 | | Model of human Anaphase-promoting complex/Cyclosome (APC15 deletion mutant), in complex with the Mitotic checkpoint complex (APC/C-CDC20-MCC) based on cryo EM data at 4.8 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Yamaguchi, M, VanderLinden, R, Dube, P, Stark, H, Schulman, B. | | Deposit date: | 2016-06-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation.
Mol.Cell, 63, 2016
|
|
5C0X
 
 | |
5CMI
 
 | | GTA mutant without mercury - E303D | | Descriptor: | Histo-blood group ABO system transferase | | Authors: | Gagnon, S.M.L, Blackler, R.J. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
5CMF
 
 | | GTA mutant with mercury - E303A | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Gagnon, S.M.L, Blackler, R.J. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
5C0W
 
 | |
