5AIO
 
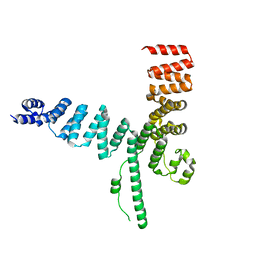 | |
7TBI
 
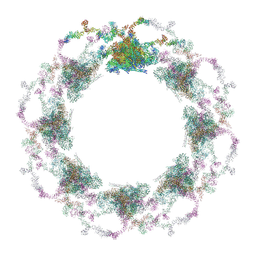 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | Descriptor: | Dyn2, Nic96 R1, Nic96 R2, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7A6H
 
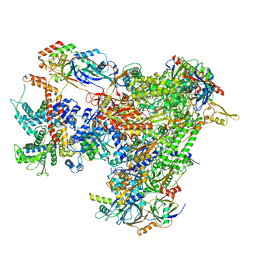 | | Cryo-EM structure of human apo RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-08-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6RQH
 
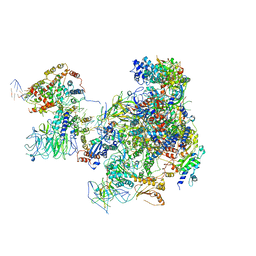 | | RNA Polymerase I Closed Conformation 1 (CC1) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-15 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
6RWE
 
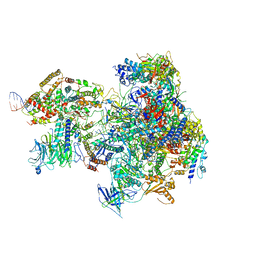 | | RNA Polymerase I Open Complex conformation 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-06-04 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
5FM7
 
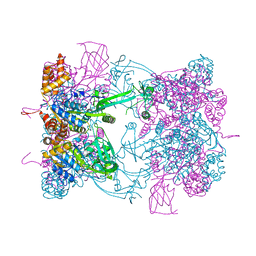 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RVB1, RVB2 | | Authors: | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
5FLV
 
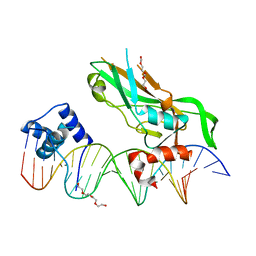 | | Crystal structure of NKX2-5 and TBX5 bound to the Nppa promoter region | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP *GP*TP*GP*AP*GP*AP*AP*G)-3', 5'-D(*TP*CP*TP*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP *TP*GP*AP*AP*GP*TP*GP*G)-3', HOMEOBOX PROTEIN NKX-2.5, ... | | Authors: | Stirnimann, C.U, Glatt, S, Mueller, C.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis.
Cell(Cambridge,Mass.), 164, 2016
|
|
5FM6
 
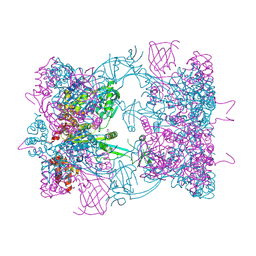 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(apo) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RVB1, ... | | Authors: | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
6TUT
 
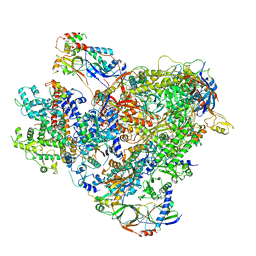 | | Cryo-EM structure of the RNA Polymerase III-Maf1 complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for RNA polymerase III transcription repression by Maf1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1K1B
 
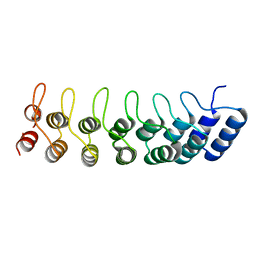 | | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family | | Descriptor: | B-cell lymphoma 3-encoded protein | | Authors: | Michel, F, Soler-Lopez, M, Petosa, C, Cramer, P, Siebenlist, U, Mueller, C.W. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family.
EMBO J., 20, 2001
|
|
1K1A
 
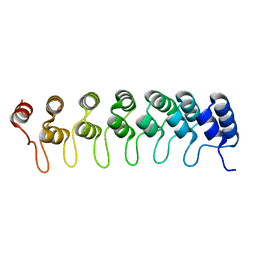 | | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family | | Descriptor: | B-cell lymphoma 3-encoded protein | | Authors: | Michel, F, Soler-Lopez, M, Petosa, C, Cramer, P, Siebenlist, U, Mueller, C.W. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family.
EMBO J., 20, 2001
|
|
5L7L
 
 | |
5L7J
 
 | |
5M2N
 
 | |
2X6U
 
 | | Crystal structure of human TBX5 in the DNA-free form | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MAGNESIUM ION, T-BOX TRANSCRIPTION FACTOR TBX5 | | Authors: | Stirnimann, C.U, Mueller, C.W. | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
2X6V
 
 | | Crystal structure of human TBX5 in the DNA-bound and DNA-free form | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5'-D(*TP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP)-3', 5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP)-3', ... | | Authors: | Ptchelkine, D, Stirnimann, C.U, Grimm, C, Mueller, C.W. | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
7OBB
 
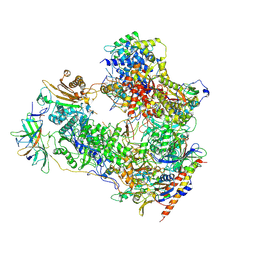 | | Cryo-EM structure of human RNA Polymerase I Open Complex | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OB9
 
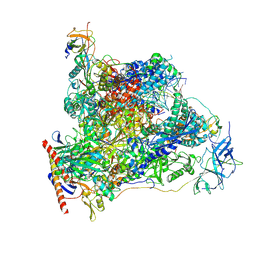 | | Cryo-EM structure of human RNA Polymerase I in elongation state | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OBA
 
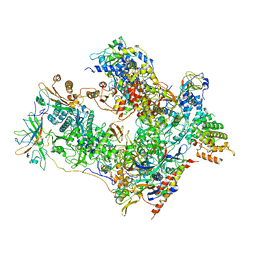 | | Cryo-EM structure of human RNA Polymerase I in complex with RRN3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OZR
 
 | |
4A8J
 
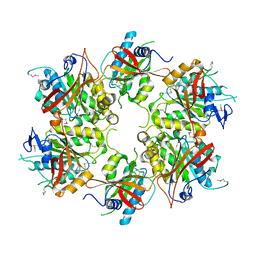 | | Crystal Structure of the Elongator subcomplex Elp456 | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2011-11-21 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Elongator Subcomplex Elp456 is a Hexameric Reca-Like ATPase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5NT7
 
 | |
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1O
 
 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
