6MEV
 
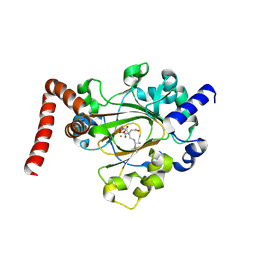 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
4WA7
 
 | | Crystal Structure of a GDP-bound Q61L Oncogenic Mutant of Human GT- Pase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-08-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
6MDB
 
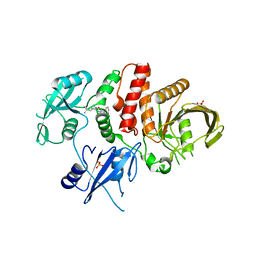 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor Pyrazolo-pyrimidinone 5 | | Descriptor: | 6-(4-amino-4-methylpiperidin-1-yl)-3-(2,3-dichlorophenyl)-5-methyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Fodor, M, Stams, T. | | Deposit date: | 2018-09-04 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | 6-Amino-3-methylpyrimidinones as Potent, Selective, and Orally Efficacious SHP2 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MDC
 
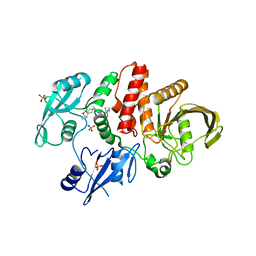 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor Pyrazolo-pyrimidinone 1 SHP389 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-[3-chloro-2-(cyclopropylamino)pyridin-4-yl]-5-methyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Fodor, M, Stams, T. | | Deposit date: | 2018-09-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of Fused Bicyclic Allosteric SHP2 Inhibitors.
J. Med. Chem., 62, 2019
|
|
5Z5T
 
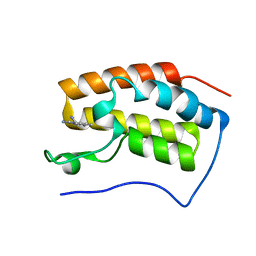 | | The first bromodomain of BRD4 with compound BDF-2141 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z5V
 
 | | The first bromodomain of BRD4 with compound BDF-1253 | | Descriptor: | Bromodomain-containing protein 4, N-{8-hydroxy-4-[(1H-imidazol-1-yl)methyl]quinolin-2-yl}acetamide | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z5U
 
 | | The first bromodomain of BRD4 with compound BDF-2254 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinoline-6,8-diol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5XFJ
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550M) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
5E6P
 
 | |
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7VLA
 
 | | Cryo-EM structure of the CCL15(27-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(27-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL9
 
 | | Cryo-EM structure of the CCL15(26-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(26-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7XA3
 
 | | Cryo-EM structure of the CCL2 bound CCR2-Gi complex | | Descriptor: | C-C motif chemokine 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7X9Y
 
 | | Cryo-EM structure of the apo CCR3-Gi complex | | Descriptor: | C-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
6JFW
 
 | | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket | | Descriptor: | PA0833-PD protein | | Authors: | Lin, X, Ye, F, Lin, S, Yang, F.L, Chen, Z.M, Cao, Y, Chen, Z.J, Gu, J, Lu, G.W. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8K4Q
 
 | |
4TQ9
 
 | | Crystal Structure of a GDP-bound G12V Oncogenic Mutant of Human GTPase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
4TQA
 
 | | Crystal Structure of a GDP-bound G13D Oncogenic Mutant of Human GTPase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
