3WQU
 
 | | Staphylococcus aureus FtsA complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
6KPM
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
7V5Q
 
 | | The dimeric structure of G80A/H81A/L137E myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Komori, H, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
7V5R
 
 | | The dimeric structure of G80A/H81A/L137D myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Komori, H, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
5GXH
 
 | | The structure of the Gemin5 WD40 domain with AAUUUUUG | | Descriptor: | GLYCEROL, Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*G)-3'), ... | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-17 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
5GXI
 
 | | Structure of the Gemin5 WD40 domain in complex with AAUUUUUGAG | | Descriptor: | Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*GP*AP*G)-3'), UNKNOWN ATOM OR ION | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
2RNN
 
 | |
2ROO
 
 | |
2RNO
 
 | |
7UOC
 
 | | Crystal structure of Orobanche minor KAI2d4 | | Descriptor: | CHLORIDE ION, KAI2d4 | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Divergent Clade KAI2 Protein in the Root Parasitic Plant Orobanche minor Is a Highly Sensitive Strigolactone Receptor and Is Involved in the Perception of Sesquiterpene Lactones.
Plant Cell.Physiol., 64, 2023
|
|
2D05
 
 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
5EWZ
 
 | | Small-molecule stabilization of the 14-3-3/Gab2 PPI interface | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, GRB2-associated-binding protein 2 | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2015-11-23 | | Release date: | 2016-05-04 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface.
Chemmedchem, 11, 2016
|
|
5EXA
 
 | | Small-molecule stabilization of the 14-3-3/Gab2 PPI interface | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, Fusicoccin A-THF derivative, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2015-11-23 | | Release date: | 2016-05-04 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface.
Chemmedchem, 11, 2016
|
|
6IRX
 
 | |
6IRY
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
7BYY
 
 | | Crystal structure of bacterial toxin | | Descriptor: | Acetyltransferase | | Authors: | Zhang, C, Yashiro, Y, Tomita, K. | | Deposit date: | 2020-04-25 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Substrate specificities of Escherichia coli ItaT that acetylates aminoacyl-tRNAs.
Nucleic Acids Res., 48, 2020
|
|
7CHD
 
 | | AtaT complexed with acetyl-methionyl-tRNAfMet | | Descriptor: | N-acetyltransferase domain-containing protein, RNA (77-MER) | | Authors: | Yashiro, Y, Tomita, K. | | Deposit date: | 2020-07-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.804 Å) | | Cite: | Mechanism of aminoacyl-tRNA acetylation by an aminoacyl-tRNA acetyltransferase AtaT from enterohemorrhagic E. coli.
Nat Commun, 11, 2020
|
|
6IRZ
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, PDX1 C-terminal-inhibiting factor 1, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6NV2
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 in complex with DP005 | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-02-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
5GHA
 
 | | Sulfur Transferase TtuA in complex with Sulfur Carrier TtuB | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Sulfur Carrier TtuB, ... | | Authors: | Chen, M, Narai, S, Tanaka, Y, Yao, M. | | Deposit date: | 2016-06-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6IRV
 
 | |
6IS0
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRW
 
 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
3NK4
 
 | |
3NK3
 
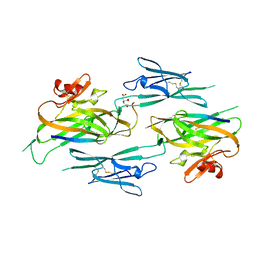 | | Crystal structure of full-length sperm receptor ZP3 at 2.6 A resolution | | Descriptor: | CITRATE ANION, Zona pellucida 3, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Monne, M, Jovine, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into Egg Coat Assembly and Egg-Sperm Interaction from the X-Ray Structure of Full-Length ZP3.
Cell(Cambridge,Mass.), 143, 2010
|
|
