5YUD
 
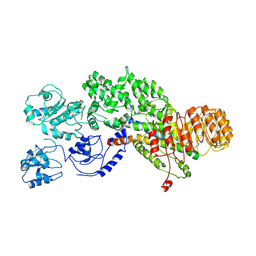 | | Flagellin derivative in complex with the NLR protein NAIP5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1e, Phase 2 flagellin,Flagellin | | Authors: | Yang, X.R, Yang, F, Wang, W.G, Lin, G.Z. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structural basis for specific flagellin recognition by the NLR protein NAIP5.
Cell Res., 28, 2018
|
|
7WMY
 
 | |
7WMZ
 
 | |
7WMW
 
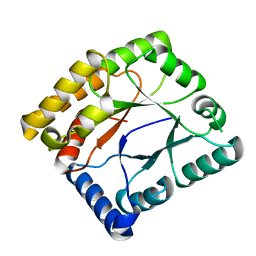 | |
7WMX
 
 | |
3BC5
 
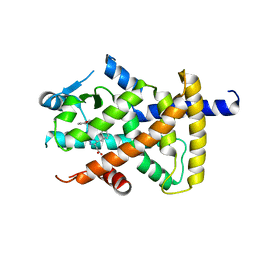 | | X-ray crystal structure of human ppar gamma with 2-(5-(3-(2-(5-methyl-2-phenyloxazol-4-yl)ethoxy)benzyl)-2-phenyl-2h-1,2,3-triazol-4-yl)acetic acid | | Descriptor: | (5-{3-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]benzyl}-2-phenyl-2H-1,2,3-triazol-4-yl)acetic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2007-11-12 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Design, synthesis and structure-activity relationships of azole acids as novel, potent dual PPAR alpha/gamma agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7E23
 
 | | SARS-CoV-2 spike in complex with the CA521 neutralizing antibody Fab (focused refinement on Fab-RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CA521 Heavy Chain, CA521 Light Chain, ... | | Authors: | Liu, C, Song, D, Dou, C. | | Deposit date: | 2021-02-04 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and function analysis of a potent human neutralizing antibody CA521 FALA against SARS-CoV-2.
Commun Biol, 4, 2021
|
|
8Y4U
 
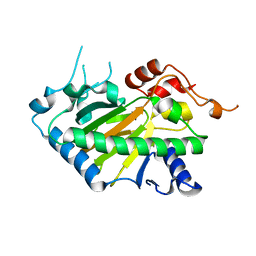 | | Crystal structure of a His1 from oryza sativa | | Descriptor: | FE (III) ION, Fe(II)/2-oxoglutarate-dependent oxygenase | | Authors: | Wang, N, Ma, J.M, Shibing, H, Beibei, Y, He, Z, Dandan, L. | | Deposit date: | 2024-01-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HPPD inhibitor sensitive protein from Oryza sativa.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
8XJ3
 
 | |
8H3T
 
 | | The crystal structure of AlpH | | Descriptor: | AlpH, GLYCEROL | | Authors: | Zhao, Y, Li, M, Jiang, M, Pan, L.F. | | Deposit date: | 2022-10-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis.
Nat Commun, 14, 2023
|
|
7WKX
 
 | | IL-17A in complex with the humanized antibody HB0017 | | Descriptor: | ACETIC ACID, Heavy chain of HB0017 Fab, Interleukin-17A, ... | | Authors: | Xu, J, Zhu, X, He, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural and functional insights into a novel pre-clinical-stage antibody targeting IL-17A for treatment of autoimmune diseases.
Int.J.Biol.Macromol., 202, 2022
|
|
3F8A
 
 | | Crystal Structure of the R132K:R111L:L121E:R59W Mutant of Cellular Retinoic Acid-Binding Protein Type II Complexed with C15-aldehyde (a retinal analog) at 1.95 Angstrom resolution. | | Descriptor: | 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue
Chempluschem, 77, 2012
|
|
3FPM
 
 | | Crystal Structure of a Squarate Inhibitor bound to MAPKAP Kinase-2 | | Descriptor: | 3-{[(1R)-1-phenylethyl]amino}-4-(pyridin-4-ylamino)cyclobut-3-ene-1,2-dione, MAP kinase-activated protein kinase 2 | | Authors: | Parris, K.D. | | Deposit date: | 2009-01-05 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification and SAR of squarate inhibitors of mitogen activated protein kinase-activated protein kinase 2 (MK-2)
Bioorg.Med.Chem., 17, 2009
|
|
3FEP
 
 | | Crystal structure of the R132K:R111L:L121E:R59W-CRABPII mutant complexed with a synthetic ligand (merocyanin) at 2.60 angstrom resolution. | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-30 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
8HRU
 
 | |
8HRI
 
 | |
8HRM
 
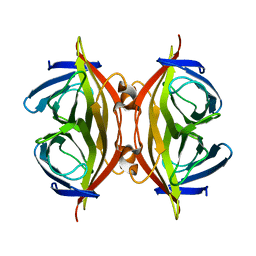 | | Cryo-EM structure of streptavidin | | Descriptor: | Streptavidin | | Authors: | Xu, J, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
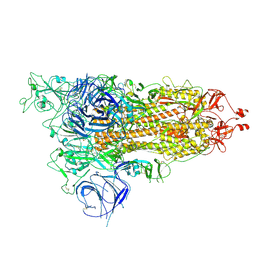
8HRK
 
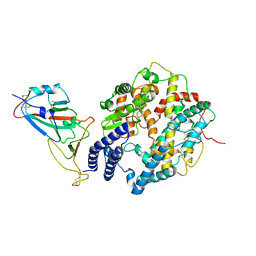 | | SARS-CoV-2 Delta S-RBD-ACE2 complex | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRN
 
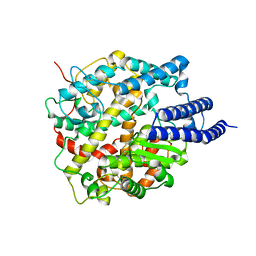 | | Cryo-EM structure of ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 | | Authors: | Xu, J, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRL
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRJ
 
 | |
3K3B
 
 | | Co-crystal structure of the human kinesin Eg5 with a novel tetrahydro-beta-carboline | | Descriptor: | 3-[(1R)-2-acetyl-6-methyl-2,3,4,9-tetrahydro-1H-beta-carbolin-1-yl]phenol, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Bussiere, D.E, Bellamacina, C, Le, V. | | Deposit date: | 2009-10-02 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The discovery of tetrahydro-beta-carbolines as inhibitors of the kinesin Eg5.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3I17
 
 | |
5X6O
 
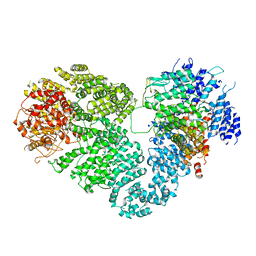 | | Intact ATR/Mec1-ATRIP/Ddc2 complex | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Wang, X, Ran, T, Cai, G. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-20 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 3.9 angstrom structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP.
Science, 358, 2017
|
|
