8GJL
 
 | | multi-drug efflux pump RE-CmeB bound with Ciprofloxacin | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GJJ
 
 | | Multi-drug efflux pump RE-CmeB Apo form | | Descriptor: | Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
4NNI
 
 | | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide | | Descriptor: | 30S ribosomal protein S1, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Yang, J, Liu, Y, Cai, Q, Lin, D. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide.
Mol.Microbiol., 95, 2015
|
|
8GK4
 
 | | Multi-drug efflux pump RE-CmeB bound with Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-17 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GK0
 
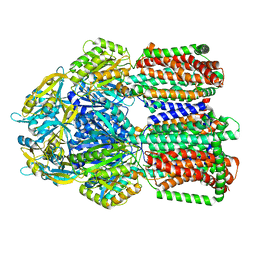 | | Multi-drug efflux pump RE-CmeB bound with Erythromycin | | Descriptor: | ERYTHROMYCIN A, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2023-03-16 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
4NNK
 
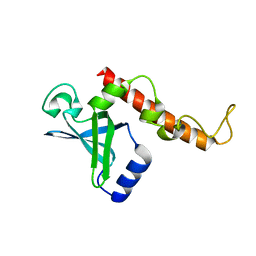 | |
3QU8
 
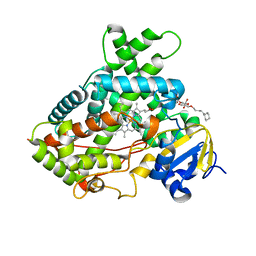 | | Crystal structure of a human cytochrome P450 2B6 (Y226H/K262R) in complex with the inhibitor 4-(4-Nitrobenzyl)pyridine. | | Descriptor: | 4-(4-nitrobenzyl)pyridine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cytochrome P450 2B6 Bound to 4-Benzylpyridine and 4-(4-Nitrobenzyl)pyridine: Insight into Inhibitor Binding and Rearrangement of Active Site Side Chains.
Mol.Pharmacol., 80, 2011
|
|
4NNH
 
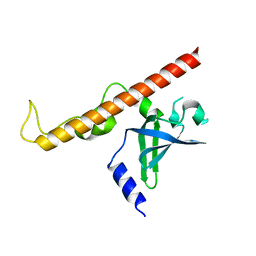 | |
7V50
 
 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V4X
 
 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V51
 
 | | BVMO_negative mutant D432V | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H.-L. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
8I60
 
 | |
3QOA
 
 | | Crystal structure of a human cytochrome P450 2B6 (Y226H/K262R) in complex with the inhibitor 4-Benzylpyridine. | | Descriptor: | 4-benzylpyridine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-02-09 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cytochrome P450 2B6 Bound to 4-Benzylpyridine and 4-(4-Nitrobenzyl)pyridine: Insight into Inhibitor Binding and Rearrangement of Active Site Side Chains.
Mol.Pharmacol., 80, 2011
|
|
7JUN
 
 | |
8IJK
 
 | | human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
7CZ6
 
 | | Protrusion structure of Omono River virus | | Descriptor: | Capsid protein | | Authors: | Shao, Q, Jia, X, Gao, Y, Liu, Z. | | Deposit date: | 2020-09-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM reveals a previously unrecognized structural protein of a dsRNA virus implicated in its extracellular transmission.
Plos Pathog., 17, 2021
|
|
7D0L
 
 | | The major capsid of Omono River virus (strain:LZ), protrusion-free status. | | Descriptor: | Capsid protein | | Authors: | Shao, Q, Jia, X, Gao, Y, Liu, Z. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM reveals a previously unrecognized structural protein of a dsRNA virus implicated in its extracellular transmission.
Plos Pathog., 17, 2021
|
|
7D0K
 
 | |
8X43
 
 | | human KCNQ2-CaM-Ebio1-S1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(4-azanyl-1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-11-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
3UAS
 
 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | Deposit date: | 2011-10-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
4NNG
 
 | |
4RAZ
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 holo-Fur | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Wang, Q, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
3HGG
 
 | | Crystal Structure of CmeR Bound to Cholic Acid | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: Crystal structures of CmeR-bile acid complexes
To be Published
|
|
7VS4
 
 | | Crystal structure of PacII_M1M2S-DNA(m6A)-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
7VRU
 
 | | Crystal structure of PacII_M1M2S-DNA-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
