4ZXU
 
 | | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289.
To be Published
|
|
5DT9
 
 | | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Erythronate-4-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Stogios, P.J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.663 Å) | | Cite: | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae
To Be Published
|
|
5HM3
 
 | | 2.25 Angstrom Resolution Crystal Structure of Long-chain-fatty-acid-AMP Ligase FadD32 from Mycobacterium tuberculosis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Warwrzak, Z, Kuhn, M.L, Shuvalova, L, Flores, K.J, Wilson, D.J, Grimes, K.D, Aldrich, C.C, Anderson, W.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-03 | | Last modified: | 2016-09-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Essential Mtb FadD32 Enzyme: A Promising Drug Target for Treating Tuberculosis.
Acs Infect Dis., 2, 2016
|
|
5HMN
 
 | | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol | | Descriptor: | AAC3-I, COENZYME A, TETRAETHYLENE GLYCOL | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol
To Be Published
|
|
5HXM
 
 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with panose | | Descriptor: | Alpha-xylosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Winsor, J, Grimshaw, S, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-31 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5I0E
 
 | | Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with isomaltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glycoside hydrolase family 31, ... | | Authors: | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5I1T
 
 | | 2.6 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Complex with Triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nocadello, S, Minasov, G, Shuvalova, L, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the SpoIID Lytic Transglycosylases Essential for Bacterial Sporulation.
J.Biol.Chem., 291, 2016
|
|
5INT
 
 | | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC | | Descriptor: | Phosphopantothenate--cysteine ligase | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC
To Be Published
|
|
5I0F
 
 | | Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with covalent intermediate | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 31, TRIETHYLENE GLYCOL, ... | | Authors: | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5I0D
 
 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with cycloalternan | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cyclic alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, ... | | Authors: | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5I82
 
 | | First Crystal Structure of E.coli Based Recombinant Diphtheria Toxin Mutant CRM197 | | Descriptor: | Diphtheria toxin, GLYCEROL, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Mishra, R.P.N, Goel, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and immunological characterization of E. coli derived recombinant CRM 197 protein used as carrier in conjugate vaccines.
Biosci.Rep., 38, 2018
|
|
5IBX
 
 | | 1.65 Angstrom Crystal Structure of Triosephosphate Isomerase (TIM) from Streptococcus pneumoniae | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.65 Angstrom Crystal Structure of Triosephosphate Isomerase (TIM) from Streptococcus pneumoniae
To Be Published
|
|
4X9K
 
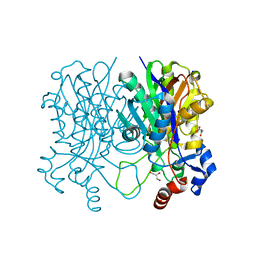 | | Beta-ketoacyl-acyl carrier protein synthase III-2 (FabH2)(C113A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Chordia, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
4YFJ
 
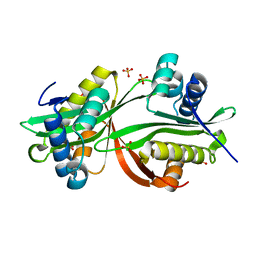 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib | | Descriptor: | Aminoglycoside 3'-N-acetyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Xu, Z, Evdokimova, E, Yim, V, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib
To Be Published
|
|
4YPI
 
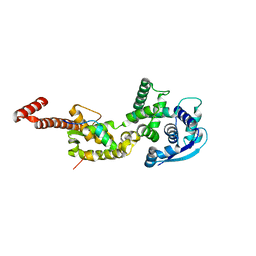 | | Structure of Ebola virus nucleoprotein N-terminal fragment bound to a peptide derived from Ebola VP35 | | Descriptor: | Nucleoprotein, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Borek, D.M, Binning, J.M, Otwinowski, Z, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.
Cell Rep, 11, 2015
|
|
4WRR
 
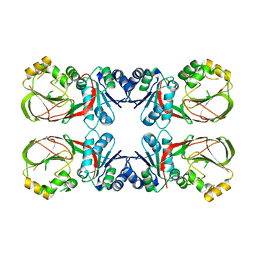 | | A putative diacylglycerol kinase from Bacillus anthracis str. Sterne | | Descriptor: | MAGNESIUM ION, putative diacylglycerol kinase | | Authors: | Hou, J, Zheng, H, Cooper, D.R, Zimmerman, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne
to be published
|
|
4WQK
 
 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, apo form | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4WQL
 
 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, kanamycin-bound | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4WZU
 
 | | Crystal structure of beta-ketoacyl-(acyl carrier protein) synthase III-2 (FabH2) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, SODIUM ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Cooper, D.R, Chordia, M.D, Zimmerman, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
To Be Published
|
|
4ZQO
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor Q67 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZQN
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor P41 | | Descriptor: | 2-chloro-N,N-dimethyl-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]benzamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZQM
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with XMP and NAD | | Descriptor: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4GKH
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4GKI
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NM-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4GFP
 
 | | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in a second conformational state | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in second conformational state
TO BE PUBLISHED
|
|
