7X2D
 
 | | Cryo-EM structure of the tavapadon-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | 1,5-dimethyl-6-[2-methyl-4-[3-(trifluoromethyl)pyridin-2-yl]oxy-phenyl]pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-15 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7X2C
 
 | | Cryo-EM structure of the fenoldopam-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7F0T
 
 | |
7F1Z
 
 | |
7F23
 
 | |
7F1O
 
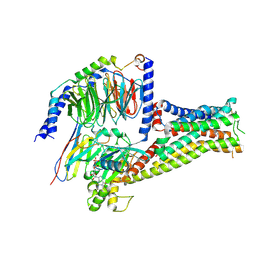 | |
7F24
 
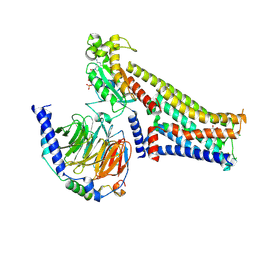 | |
7XJL
 
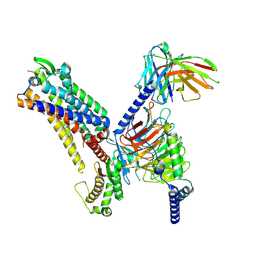 | | Cryo-EM structure of the spexin-bound GALR2-miniGq complex | | Descriptor: | Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XJK
 
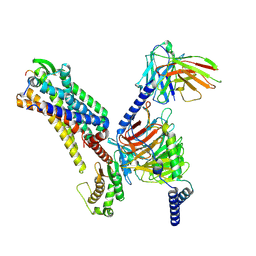 | |
7XJJ
 
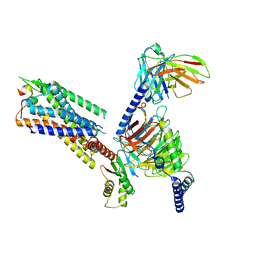 | | Cryo-EM structure of the galanin-bound GALR1-miniGo complex | | Descriptor: | G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Galanin, Galanin receptor type 1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2HV9
 
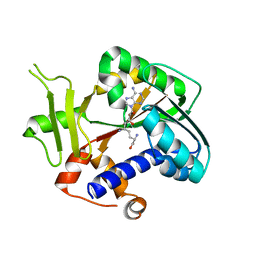 | | Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase in complex with sinefungin | | Descriptor: | SINEFUNGIN, mRNA cap guanine-N7 methyltransferase | | Authors: | Lima, C.D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Analysis of Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase, Structure of the Enzyme Bound to Sinefungin, and Evidence That Cap Methyltransferase Is the Target of Sinefungin's Antifungal Activity
J.Biol.Chem., 281, 2006
|
|
6IP0
 
 | |
6IP4
 
 | |
2KHP
 
 | |
3L0L
 
 | |
5WYK
 
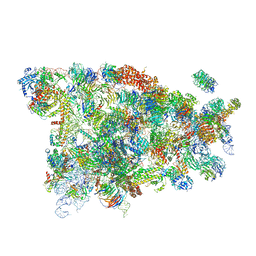 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2017-05-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WYJ
 
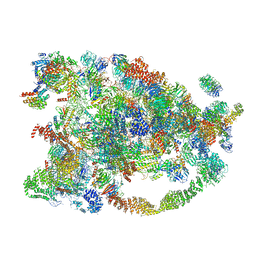 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WY3
 
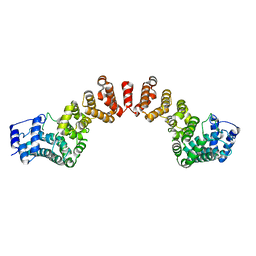 | |
7T86
 
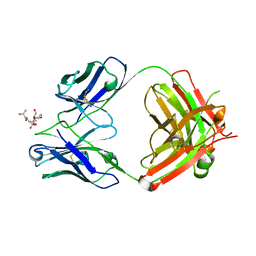 | | Crystal Structure of Fab CR5133 / Phospho-SD Peptide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab CR5133 Heavy Chain, Antibody Fab CR5133 Light Chain, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
7T82
 
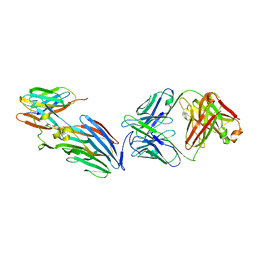 | | Crystal Structure of LEUKOCIDIN E/CENTYRIN S26/FAB B438 | | Descriptor: | Antibody Fab Heavy Chain, Antibody Fab Light Chain, Centyrin S26, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
7T87
 
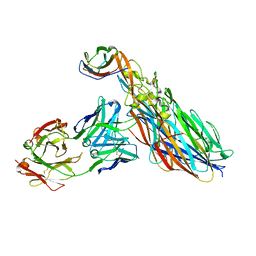 | | CRYSTAL STRUCTURE OF LEUKOCIDIN AB/CENTYRIN S17/FAB 214F COMPLEX | | Descriptor: | Antibody Fab B214 Heavy Chain, Antibody Fab B214 Light Chain, Centyrin S17, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
1Z3C
 
 | | Encephalitozooan cuniculi mRNA Cap (Guanine-N7) Methyltransferasein complexed with AzoAdoMet | | Descriptor: | S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE, mRNA CAPPING ENZYME | | Authors: | Hausmann, S, Zhang, S, Fabrega, C, Schneller, S.W, Lima, C.D, Shuman, S. | | Deposit date: | 2005-03-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Encephalitozoon cuniculi mRNA cap (guanine N-7) methyltransferase: methyl acceptor specificity, inhibition BY S-adenosylmethionine analogs, and structure-guided mutational analysis.
J.Biol.Chem., 280, 2005
|
|
5EIB
 
 | |
7BTN
 
 | |
2Y78
 
 | | Crystal structure of BPSS1823, a Mip-like chaperone from Burkholderia pseudomallei | | Descriptor: | CHLORIDE ION, GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Norville, I.H, O'Shea, K, Sarkar-Tyson, M, Harmer, N.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | The Structure of a Burkholderia Pseudomallei Immunophilin-Inhibitor Complex Reveals New Approaches to Antimicrobial Development
Biochem.J., 437, 2011
|
|
