7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | Authors: | Xiao, J, Zhu, Q, Wang, G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CHE
 
 | |
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
3NTE
 
 | |
7EY5
 
 | |
7EZV
 
 | |
7EY4
 
 | | Local CryoEM of the SARS-CoV-2 S6PV2 in complex with BD-667 | | Descriptor: | BD-667 H, BD-667 L, Spike glycoprotein, ... | | Authors: | Liu, P.L. | | Deposit date: | 2021-05-29 | | Release date: | 2021-09-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants.
Cell Res., 31, 2021
|
|
5TX4
 
 | | Derivative of mouse TGF-beta2, with a deletion of residues 52-71 and K25R, R26K, L51R, A74K, C77S, L89V, I92V, K94R T95K, I98V single amino acid substitutions, bound to human TGF-beta type II receptor ectodomain residues 15-130 | | Descriptor: | TGF-beta receptor type-2, Transforming growth factor beta-2 | | Authors: | Hinck, A.P, Kim, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | An engineered transforming growth factor beta (TGF-beta ) monomer that functions as a dominant negative to block TGF-beta signaling.
J. Biol. Chem., 292, 2017
|
|
3MBS
 
 | | Crystal structure of 8mer PNA | | Descriptor: | 1,2-ETHANEDIOL, Peptide Nucleic Acid | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
5TX6
 
 | |
5TX2
 
 | | Miniature TGF-beta2 3-mutant monomer | | Descriptor: | Transforming growth factor beta-2 | | Authors: | Taylor, A.B, Kim, S.K, Hart, P.J, Hinck, A.P. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An engineered transforming growth factor beta (TGF-beta ) monomer that functions as a dominant negative to block TGF-beta signaling.
J. Biol. Chem., 292, 2017
|
|
4Z3D
 
 | |
7Y0V
 
 | | The co-crystal structure of BA.1-RBD with Fab-5549 | | Descriptor: | 5549-Fab, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2022-06-06 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
7Y0C
 
 | | Crystal structure of BD55-1403 and SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-1403 Fab heavy chain, BD55-1403 Fab light chain, ... | | Authors: | Zhang, Z, Xiao, J. | | Deposit date: | 2022-06-04 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
7VNE
 
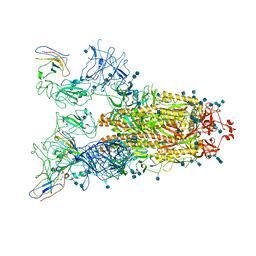 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113.1 (UUU-state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VND
 
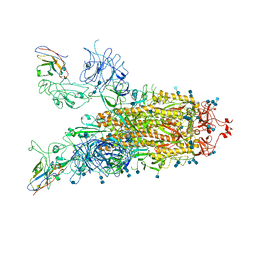 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UUD-state, state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNB
 
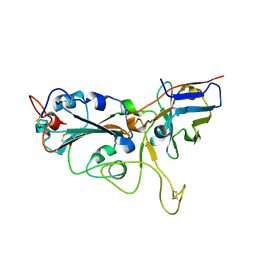 | | Crystal structure of the SARS-CoV-2 RBD in complex with a human single domain antibody n3113 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, n3113 | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNC
 
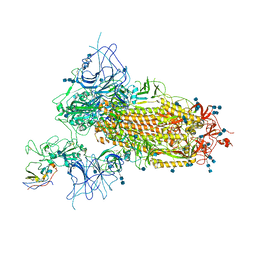 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UDD-state, state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
7WHK
 
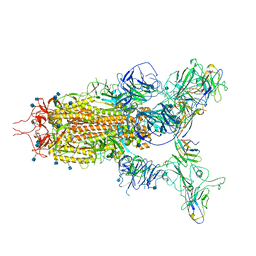 | | The state 3 complex structure of Omicron spike with Bn03 (2-up RBD, 5 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHI
 
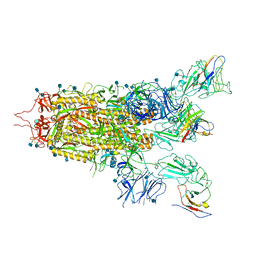 | | The state 2 complex structure of Omicron spike with Bn03 (2-up RBD, 4 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHJ
 
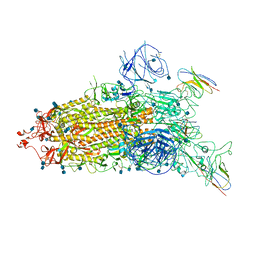 | | The state 1 complex structure of Omicron spike with Bn03 (1-up RBD, 3 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7VIC
 
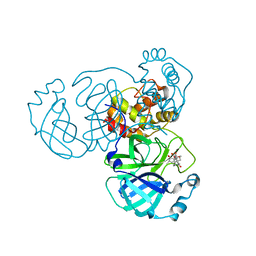 | | The crystal structure of SARS-CoV-2 3C-like protease in complex with a traditional Chinese Medicine Inhibitors | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 3C-like proteinase | | Authors: | Zhong, B, Chen, B, Zhou, H, Sun, L. | | Deposit date: | 2021-09-26 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oridonin Inhibits SARS-CoV-2 by Targeting Its 3C-Like Protease.
Small Sci, 2, 2022
|
|
7WRO
 
 | | Local structure of BD55-3372 and delta spike | | Descriptor: | 3372H, 3372L, Spike protein S1 | | Authors: | Liu, P.L. | | Deposit date: | 2022-01-27 | | Release date: | 2022-06-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7WRL
 
 | |
