3U1S
 
 | | Crystal structure of human Fab PGT145, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Fab PGT145 Heavy chain, Fab PGT145 Light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Diwanji, D, Burton, D.R, Wilson, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U46
 
 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
8IYQ
 
 | | Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-04-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8HEC
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HEB
 
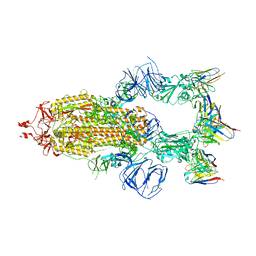 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
6LY5
 
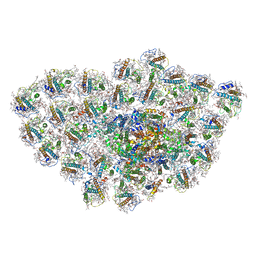 | | Organization and energy transfer in a huge diatom PSI-FCPI supercomplex | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Xiong, P, Caizhe, X. | | Deposit date: | 2020-02-13 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structural basis for energy transfer in a huge diatom PSI-FCPI supercomplex.
Nat Commun, 11, 2020
|
|
8T5C
 
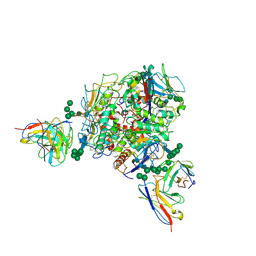 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
7BWQ
 
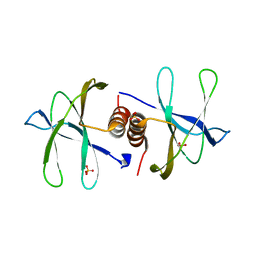 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
6AJH
 
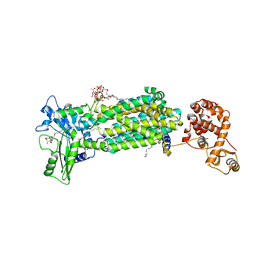 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with AU1235 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1-(2-adamantyl)-3-[2,3,4-tris(fluoranyl)phenyl]urea, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJI
 
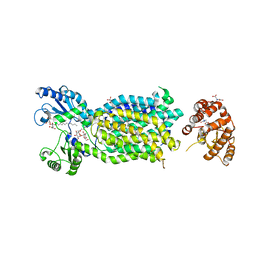 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with Rimonabant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJF
 
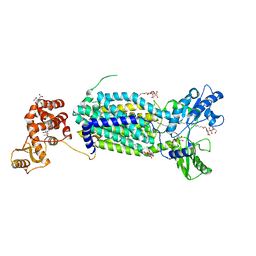 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Drug exporters of the RND superfamily-like protein,Endolysin, alpha-D-glucopyranosyl 6-O-dodecyl-alpha-D-glucopyranoside | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJG
 
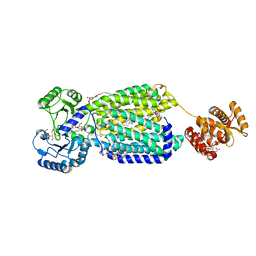 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SQ109 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, DODECYL-BETA-D-MALTOSIDE, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJJ
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with ICA38 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4,6-difluoro-N-(spiro[5.5]undecan-3-yl)-1H-indole-2-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
3KEY
 
 | | Crystal structure of S. cerevisiae Stn1 C-terminal | | Descriptor: | Protein STN1 | | Authors: | Sun, J, Yu, E.Y, Yang, Y.T, Confer, L.A, Sun, S.H, Wan, K, Lue, N.F, Lei, M. | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres.
Genes Dev., 23, 2009
|
|
3KF8
 
 | | Crystal structure of C. tropicalis Stn1-ten1 complex | | Descriptor: | Protein stn1, Protein ten1 | | Authors: | Sun, J, Yu, E.Y, Yang, Y.T, Confer, L.A, Sun, S.H, Wan, K, Lue, N.F, Lei, M. | | Deposit date: | 2009-10-27 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres.
Genes Dev., 23, 2009
|
|
5KZI
 
 | | Crystal structure of human Pim-1 kinase in complex with an imidazopyridazine inhibitor. | | Descriptor: | Serine/threonine-protein kinase pim-1, ~{N}-[4-[(3~{S})-3-azanylpiperidin-1-yl]pyridin-3-yl]-2-[2,6-bis(fluoranyl)phenyl]imidazo[1,5-b]pyridazin-7-amine | | Authors: | Mohr, C. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of imidazopyridazines as potent Pim-1/2 kinase inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
3WUB
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUG
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUF
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
4OM1
 
 | | Crystal structure of antibody VRC07-I30Q, G54W, S58N in complex with clade A/E 93TH057 HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen binding fragment of heavy chain: Antibody VRC01, Antigen binding fragment of light chain: Antibody VRC01, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2014-01-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.131 Å) | | Cite: | Enhanced Potency of a Broadly Neutralizing HIV-1 Antibody In Vitro Improves Protection against Lentiviral Infection In Vivo.
J.Virol., 88, 2014
|
|
4OLZ
 
 | |
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
4OM0
 
 | |
