5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | Descriptor: | Major capsid protein, UL17, UL25, ... | | Authors: | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
4PJW
 
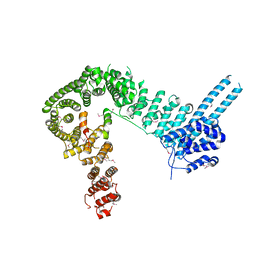 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1), with bound MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PK7
 
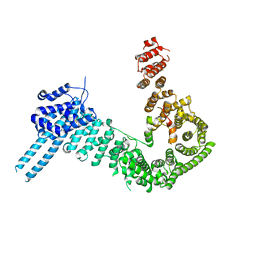 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) with bound MES, native proteins | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PJU
 
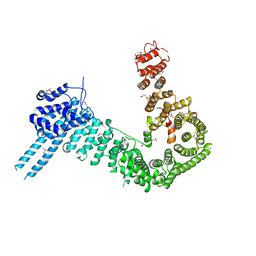 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1V0J
 
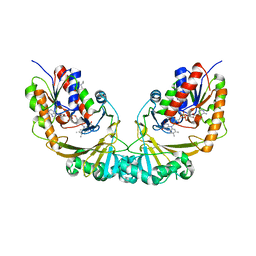 | | Udp-galactopyranose mutase from Mycobacterium tuberculosis | | Descriptor: | BICINE, FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Naismith, J.H. | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state.
J. Mol. Biol., 348, 2005
|
|
2F1G
 
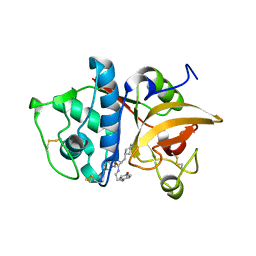 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3IF1
 
 | | Crystal structure of 237mAb in complex with a GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Immunoglobulin heavy chain (IgG2a), Immunoglobulin light chain (IgG2a), ... | | Authors: | Brooks, C.L, Evans, S.V, Borisova, S.N. | | Deposit date: | 2009-07-23 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Antibody recognition of a unique tumor-specific glycopeptide antigen.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IET
 
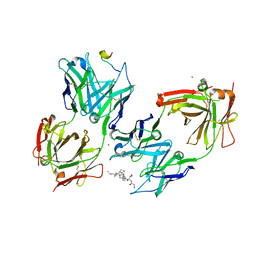 | | Crystal Structure of 237mAb with antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Immunoglobulin heavy chain (IgG2a), Immunoglobulin light chain (IgG2a), ... | | Authors: | Brooks, C.L, Evans, S.V, Borisova, S.N. | | Deposit date: | 2009-07-23 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody recognition of a unique tumor-specific glycopeptide antigen.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7F9W
 
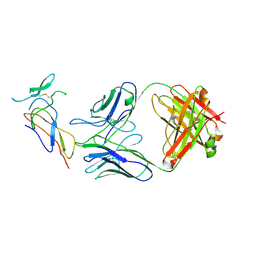 | | CD25 in complex with Fab | | Descriptor: | Heavy chain of Fab, Interleukin-2 receptor subunit alpha, Light chain of Fab | | Authors: | Liu, C. | | Deposit date: | 2021-07-05 | | Release date: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two novel human anti-CD25 antibodies with antitumor activity inversely related to their affinity and in vitro activity.
Sci Rep, 11, 2021
|
|
4EXA
 
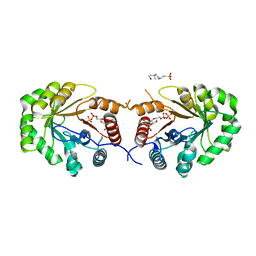 | | Crystal structure of the PA4992, the putative aldo-keto reductase from Pseudomona aeruginosa | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein | | Authors: | Sandalova, T, Schnell, R, Schneider, G. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ETR
 
 | | X-ray structure of PA2169 from Pseudomonas aeruginosa | | Descriptor: | Putative uncharacterized protein | | Authors: | Schnell, R, Sandalova, T, Lindqvist, Y, Schneider, G. | | Deposit date: | 2012-04-24 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4EXB
 
 | |
4ES6
 
 | |
5TDK
 
 | | RNA decamer duplex with eight 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
7F7E
 
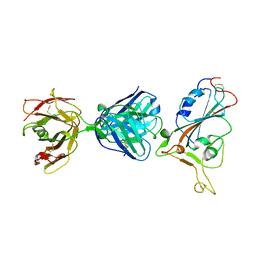 | | SARS-CoV-2 S protein RBD in complex with A5-10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A5-10 Fab, Light chain of A5-10 Fab, ... | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants.
Emerg Microbes Infect, 11, 2022
|
|
2JVN
 
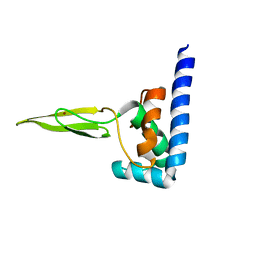 | | Domain C of human PARP-1 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Hoffman, D, Tao, Z, Liu, H. | | Deposit date: | 2007-09-24 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif.
Biochemistry, 47, 2008
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
5TDJ
 
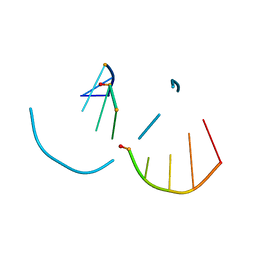 | | RNA decamer duplex with four 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
7EY9
 
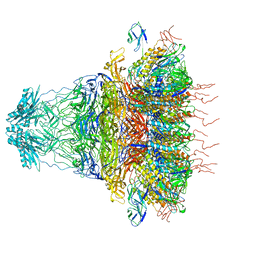 | | tail proteins | | Descriptor: | Tail fiber protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY6
 
 | | The portal protein (GP8) of bacteriophage T7 | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EYB
 
 | | core proteins | | Descriptor: | Internal virion protein gp14, Internal virion protein gp15, Peptidoglycan transglycosylase gp16 | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY7
 
 | | bacteriophage T7 tail complex | | Descriptor: | Internal virion protein gp14, Tail fiber protein, Tail tubular protein gp11, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY8
 
 | | portal | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5VT2
 
 | | Crystal structure of growth differentiation factor | | Descriptor: | 1,2-ETHANEDIOL, Growth/differentiation factor 15 | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2017-05-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Long-acting MIC-1/GDF15 molecules to treat obesity: Evidence from mice to monkeys.
Sci Transl Med, 9, 2017
|
|
