8IQF
 
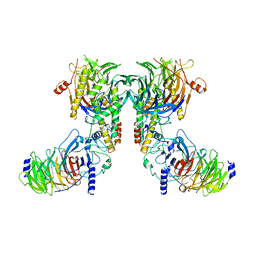 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQG
 
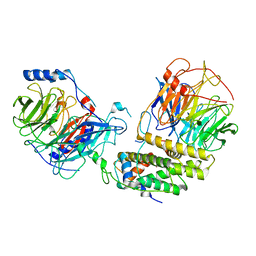 | | Cryo-EM structure of the monomeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
4JII
 
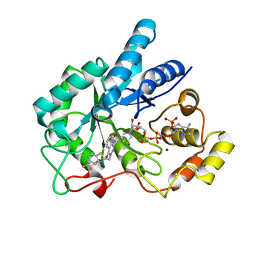 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
5XP3
 
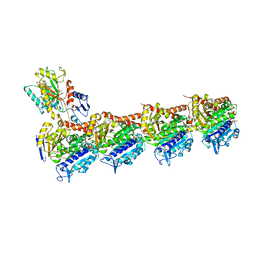 | | Crystal structure of apo T2R-TTL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
7OVP
 
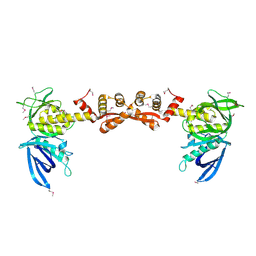 | |
7OD9
 
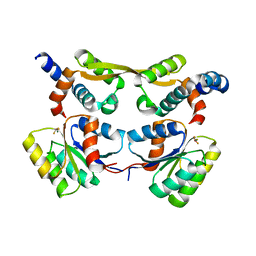 | | Crystal structure of activated CheY fused to the C-terminal domain of CheF | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, C-terminal domain of CheF from Methanococcus maripaludis, MAGNESIUM ION, ... | | Authors: | Altegoer, F, Weiland, P, Bange, G. | | Deposit date: | 2021-04-29 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of archaellar rotational switching.
Nat Commun, 13, 2022
|
|
1NW3
 
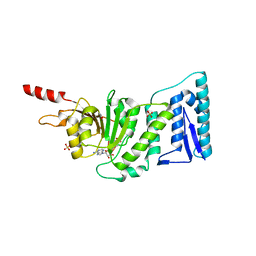 | | Structure of the Catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Min, J.R, Feng, Q, Li, Z.H, Zhang, Y, Xu, R.M. | | Deposit date: | 2003-02-05 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase
Cell(Cambridge,Mass.), 112, 2003
|
|
6OWN
 
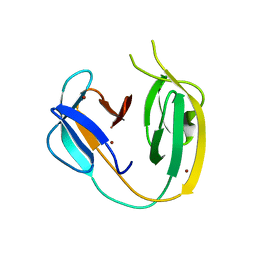 | |
6QFY
 
 | | CRYSTAL STRUCTURE OF PORCINE HEMAGGLUTINATING ENCEPHALOMYELITIS VIRUS SPIKE PROTEIN LECTIN DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Huizinga, E.G, Bakkers, M, Lang, Y. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8K9D
 
 | | Structure of human Caprin-2 HR1 domain | | Descriptor: | Caprin-2 | | Authors: | Song, X.M. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the Caprin-2 HR1 domain in canonical Wnt signaling.
J.Biol.Chem., 300, 2024
|
|
9LSM
 
 | | The crystal structure of PDE5A with L9 | | Descriptor: | 1-[(13~{S},15~{R})-4-bromanyl-15-(3-chloranyl-4-methoxy-phenyl)-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaen-12-yl]ethanone, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
9LSL
 
 | | The crystal structure of PDE5A with L1 | | Descriptor: | (13~{S},15~{R})-15-(3-chloranyl-4-methoxy-phenyl)-12-ethanoyl-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaene-4-carbonitrile, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
8H3S
 
 | | Substrate-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain, Serine protease 1 | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
8H3U
 
 | | Inhibitor-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
8HFY
 
 | | SARS-CoV-2 Omicron BA.1 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFX
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HG0
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFZ
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
6K9N
 
 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
8IBU
 
 | | Cryo-EM structure of the erythromycin-bound motilin receptor-Gq protein complex | | Descriptor: | ERYTHROMYCIN A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | You, C, Jiang, Y, Xu, H.E, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
8IBV
 
 | | Cryo-EM structure of the motilin-bound motilin receptor-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Jiang, Y, Xu, H.E, You, C, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
5XS5
 
 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
6K9P
 
 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6KBE
 
 | | Structure of Deubiquitinase | | Descriptor: | Polyubiquitin-C, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6LQC
 
 | | Crystal structure of Cyclohexylamine Oxidase from Erythrobacteraceae bacterium | | Descriptor: | Cyclohexylamine Oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Huang, Z.D. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Asymmetric Synthesis of a Key Dextromethorphan Intermediate and Its Analogues Enabled by a New Cyclohexylamine Oxidase: Enzyme Discovery, Reaction Development, and Mechanistic Insight.
J.Org.Chem., 85, 2020
|
|
