7X0O
 
 | |
7X0Q
 
 | |
7X0R
 
 | |
7X0I
 
 | |
7X0N
 
 | |
7X0M
 
 | |
7X0J
 
 | |
7XDD
 
 | | Cryo-EM structure of EDS1 and PAD4 | | Descriptor: | Lipase-like PAD4, Protein EDS1 | | Authors: | Huang, S.J, Jia, A.L, Sun, Y, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-03-26 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
7DY7
 
 | | Discovery of Novel Small-molecule Inhibitors of PD-1/PD-L1 Axis that Promotes PD-L1 Internalization and Degradation | | Descriptor: | 2-[[3-[[5-(2-methyl-3-phenyl-phenyl)-1,3,4-oxadiazol-2-yl]amino]phenyl]methylamino]ethanol, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Wang, T.Y, Lu, M.L, Jiang, S, Xiao, Y.B. | | Deposit date: | 2021-01-20 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of the PD-1/PD-L1 Axis That Promote PD-L1 Internalization and Degradation.
J.Med.Chem., 65, 2022
|
|
7E58
 
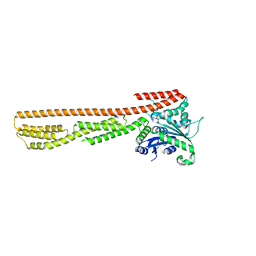 | | interferon-inducible anti-viral protein 2 | | Descriptor: | Guanylate-binding protein 2 | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E59
 
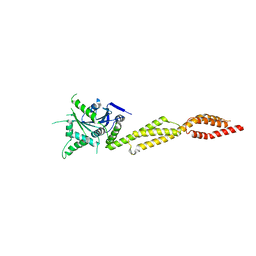 | | interferon-inducible anti-viral protein truncated | | Descriptor: | Guanylate-binding protein 5 | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E5A
 
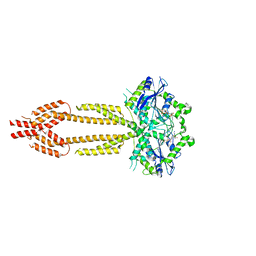 | | interferon-inducible anti-viral protein R356A | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FI4
 
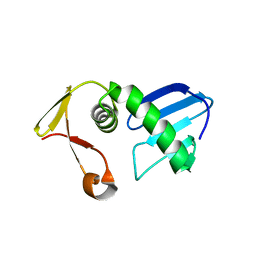 | | Structure of AcrIF13 | | Descriptor: | AcrIF13 | | Authors: | Feng, Y, Gao, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mechanistic insights into the inhibition of the CRISPR-Cas surveillance complex by anti-CRISPR protein AcrIF13.
J.Biol.Chem., 298, 2022
|
|
4LU5
 
 | |
4LQF
 
 | | Structure of murine IgG2b A2C7-Fab in complex with vaccinia antigen A33R at the resolution of 2.3 Angstroms | | Descriptor: | A33R, Murine IgG2b A2C7 Heavy chain Fab domain, Murine IgG2b A2C7 Light chain Fab domain, ... | | Authors: | Matho, M.H, Schlossman, A.M, Zajonc, D.M. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Anti-A33 Antibodies Reveal a Potent Cross-Species Orthopoxviruses Neutralizer.
Plos Pathog., 11, 2015
|
|
7W2B
 
 | |
7W2C
 
 | | The closed conformation of the sigma-1 receptor from Xenopus laevis complexed with PRE084 | | Descriptor: | 2-morpholin-4-ylethyl 1-phenylcyclohexane-1-carboxylate, GOLD ION, Sigma non-opioid intracellular receptor 1, ... | | Authors: | Meng, F, Sun, Z, Zhou, X. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | An open-like conformation of the sigma-1 receptor reveals its ligand entry pathway.
Nat Commun, 13, 2022
|
|
7W2E
 
 | |
7W2G
 
 | |
7W2H
 
 | |
7W2D
 
 | |
7W2F
 
 | |
7WKU
 
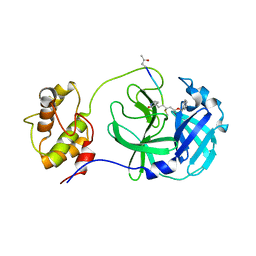 | | Structure of PDCoV Mpro in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Peptidase C30 | | Authors: | Wang, F.H, Yang, H.T. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Porcine Deltacoronavirus Main Protease Reveals a Conserved Target for the Design of Antivirals.
Viruses, 14, 2022
|
|
7ELM
 
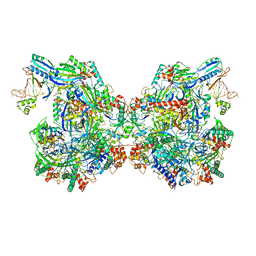 | | Structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7ELN
 
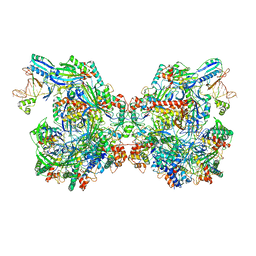 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | 54-MER DNA, AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
