8B3K
 
 | |
8BF4
 
 | | Crystal structure of Mouse Plexin-B1 (20-535) in complex with VHH15 and VHH14 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Cowan, R, Hall, G, Carr, M. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nanobody inhibitors of Plexin-B1 identify allostery in plexin-semaphorin interactions and signaling.
J.Biol.Chem., 299, 2023
|
|
5N9B
 
 | | Crystal Structure of unliganded human IL-17RA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A | | Authors: | Rondeau, J.-M, Goepfert, A. | | Deposit date: | 2017-02-24 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IL-17-induced dimerization of IL-17RA drives the formation of the IL-17 signalosome to potentiate signaling.
Cell Rep, 41, 2022
|
|
6HGA
 
 | |
7ZCA
 
 | | Crystal structure of the F191M/F201A variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with benzyl phenyl sulfoxide | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative dehydrogenase/oxygenase subunit (Flavoprotein), ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-26 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5NAN
 
 | | Crystal Structure of human IL-17AF in complex with human IL-17RA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Rondeau, J.-M, Goepfert, A. | | Deposit date: | 2017-02-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The human IL-17A/F heterodimer: a two-faced cytokine with unique receptor recognition properties.
Sci Rep, 7, 2017
|
|
5C4K
 
 | | APH(2")-IVa in complex with GET (G418) at room temperature | | Descriptor: | APH(2'')-Id, GENETICIN | | Authors: | Kaplan, E, Guichou, J.F, Berrou, K, Chaloin, L, Leban, N, Lallemand, P, Barman, T, Serpersu, E.H, Lionne, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Aminoglycoside binding and catalysis specificity of aminoglycoside 2-phosphotransferase IVa: A thermodynamic, structural and kinetic study.
Biochim.Biophys.Acta, 1860, 2016
|
|
6HG4
 
 | |
5C4L
 
 | | Conformational alternate of sisomicin in complex with APH(2")-IVa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, (2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-ol, APH(2'')-Id | | Authors: | Kaplan, E, Guichou, J.F, Berrou, K, Chaloin, L, Leban, N, Lallemand, P, Barman, T, Serpersu, E.H, Lionne, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aminoglycoside binding and catalysis specificity of aminoglycoside 2-phosphotransferase IVa: A thermodynamic, structural and kinetic study.
Biochim.Biophys.Acta, 1860, 2016
|
|
8HI9
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with Robinetin | | Descriptor: | 3,7-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-11-19 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of Polyphenolic Natural Products as SARS-CoV-2 M pro Inhibitors for COVID-19.
Pharmaceuticals, 16, 2023
|
|
6WAQ
 
 | |
6WAR
 
 | |
4EQQ
 
 | | Structure of Ltp, a superinfection exclusion protein from the Streptococcus thermophilus temperate phage TP-J34 | | Descriptor: | PHOSPHATE ION, Putative host cell surface-exposed lipoprotein, SULFATE ION | | Authors: | Bebeacua, C, Lorenzo, C, Blangy, S, Spinelli, S, Heller, K, Cambillau, C. | | Deposit date: | 2012-04-19 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of a superinfection exclusion lipoprotein from phage TP-J34 and identification of the tape measure protein as its target.
Mol.Microbiol., 89, 2013
|
|
7T2Q
 
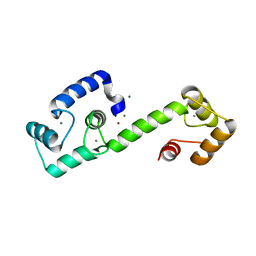 | | PEGylated Calmodulin-1 (K148U) | | Descriptor: | CALCIUM ION, Calmodulin-1, MAGNESIUM ION, ... | | Authors: | Mackay, J.P, Payne, R.J, Patel, K, Dowman, L.J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Site-selective photocatalytic functionalization of peptides and proteins at selenocysteine.
Nat Commun, 13, 2022
|
|
5NHW
 
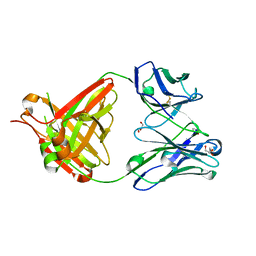 | | CRYSTAL STRUCTURE OF THE BIMAGRUMAB Fab | | Descriptor: | GLYCEROL, anti-human ActRII Bimagrumab Fab heavy-chain, anti-human ActRII Bimagrumab Fab light-chain | | Authors: | Rondeau, J.-M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Blockade of activin type II receptors with a dual anti-ActRIIA/IIB antibody is critical to promote maximal skeletal muscle hypertrophy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
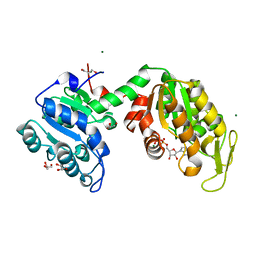
3ZLB
 
 | |
3ZG1
 
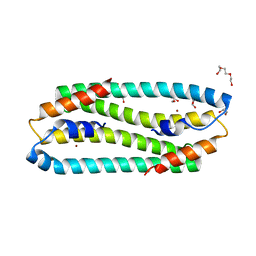 | | NI-BOUND FORM OF M123A MUTANT OF CUPRIAVIDUS METALLIDURANS CH34 CNRXS | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Girard, E, Coves, J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal Sensing and Signal Transduction by Cnrx from Cupriavidus Metallidurans Ch34: Role of the Only Methionine Assessed by a Functional, Spectroscopic, and Theoretical Study
Metallomics, 6, 2014
|
|
5NHR
 
 | |
7ZAN
 
 | |
7Z94
 
 | | Crystal structure of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with indole | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, INDOLE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7Z99
 
 | |
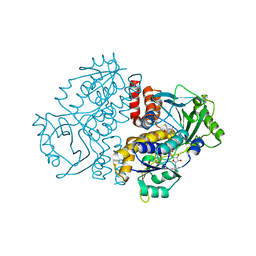
7Z4X
 
 | |
7Z98
 
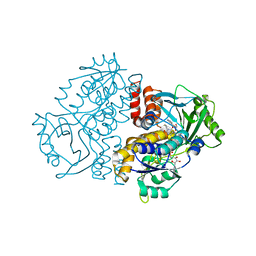 | | Crystal structure of F191M variant Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with methyl phenyl sulfide | | Descriptor: | (methylsulfanyl)benzene, 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7Z97
 
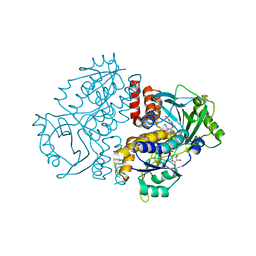 | | Crystal structure of the F191M variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with 6-bromoindole | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7V90
 
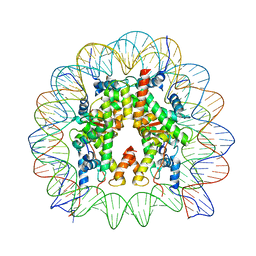 | | Telomeric mononucleosome | | Descriptor: | DNA (145-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
